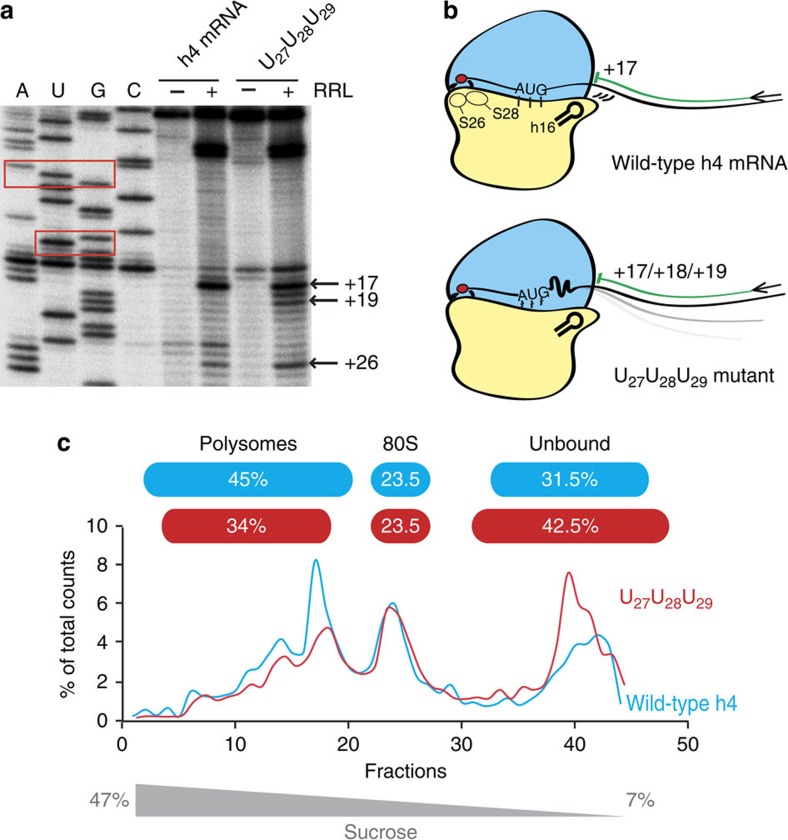
Figure 2. Ribosome toe-prints and polysome fractionation with h4 mRNA and mutant.
(a) Initiation complexes were assembled in RRL extracts in the presence of cycloheximide and hygromycin B to stall the initiation complexes on the AUG codon. Reaction samples were separated on 8% denaturing PAGE together with the appropriate sequencing ladder (shown on the left). AUG initiation codon and GUG codon are boxed. Toe-print positions were numbered starting on the A of the AUG codon (+17 position corresponds to h4 nt 27). (b) Model of h4 interaction within the 80S ribosomal particle. Accurate positioning of h4 mRNA results from interactions with helix h16 from 18S rRNA. Mutation of nts 27–29 induces toe-print shifts to +18 and +19 indicating that the mRNA is not accurately maintained into the mRNA channel. (c) Polysome fractionation of translation extracts programmed with wild-type h4 mRNA and derived triple mutant. Ribosome assembly and translation was studied in RRL programmed with 5′-end radiolabelled m7G-capped h4 mRNA. Unblocked translation extracts were separated on 7–47% sucrose gradients and radiolabelled mRNAs were detected by Cerenkov counting. The graph represents the radioactivity in the different fractions expressed as a percentage of the total radioactive counts. The positions of polysomes, 80S and free mRNA are indicated. The sums of counts measured in polysomes, 80S particles and not assembled (unbound) are indicated in the blue and red bars for wild-type and triple mutant, respectively.