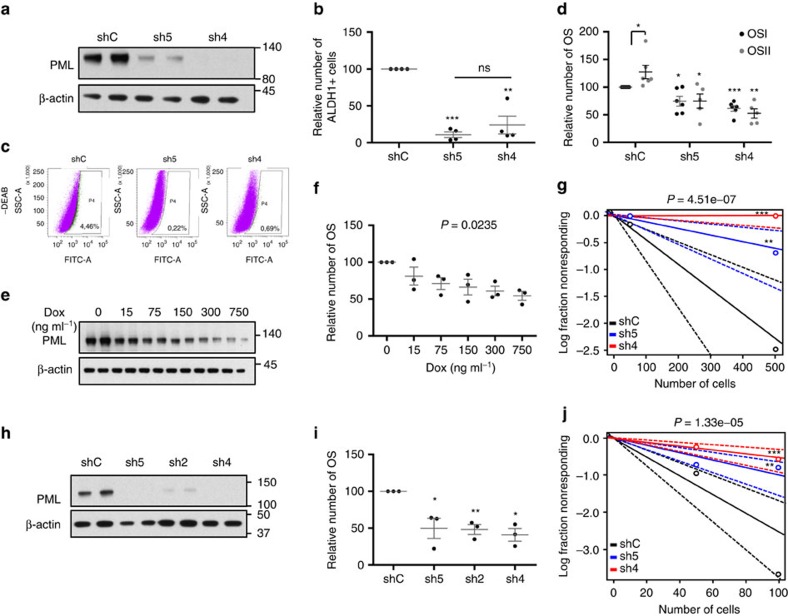
Figure 1. Genetic targeting of PML hampers breast cancer initiation potential.
(a) PML levels (representative western blot out of four independent experiments) upon PML silencing with two shRNAs (sh) in MDA-MB-231 cells. (b) Percentage of ALDH1+ cells upon PML silencing with two shRNAs in MDA-MB-231 cells (n=4). (c) Representative flow cytometry analysis out of three independent experiments of the ALDH1+ population in shC or shPML-transduced MDA-MB-231 cells (FITC: fluorescein-isothiocyanate, SSC-A: side-scatter). (d) Effect of PML silencing on primary (OSI) and secondary (OSII) OS formation in MDA-MB-231 cells (n=5 for OSII in shPML cells and n=6 for shC and OSI in shPML cells). (e,f) PML levels (representative western blot out of three independent experiments) (e) and OS formation (n=3) (f) upon PML inducible silencing (shPML#4) with the indicated doses of doxycycline in MDA-MB-231 cells. (g) Limiting dilution experiment after xenotransplantation. Nude mice were inoculated with 500,000 or 50,000 MDA-MB-231 cells (n=12 injections per experimental condition). Tumour-initiating cell number was calculated using the ELDA platform. A log-fraction plot of the limiting dilution model fitted to the data is presented. The slope of the line is the log-active cell fraction (solid lines: mean; dotted lines: 95% confidence interval; circles: values obtained in each cell dilution). (h) PML levels (representative western blot out of four independent experiments) upon PML silencing in the PDX44-derived cell line. (i) OSI formation upon PML silencing in PDX44 cells (n=3). (j) Limiting dilution experiment after xenotransplantation. Nude mice were inoculated either with 100,000 or 10,000 PDX44 cells (n=20 injections per experimental condition). Tumour-initiating cell number was calculated using the ELDA platform as in g. Error bars represent s.e.m., P value (*P<0.05; **P<0.01; ***P<0.001 compared with shC or as indicated). Statistics test: one-tail unpaired t-test (b,d,i), analysis of variance (f) and χ2-test (g,j). dox, doxycycline; OS, oncospheres; shC, Scramble shRNA; sh2, sh4 and sh5, shRNA against PML.