FIG 4 .
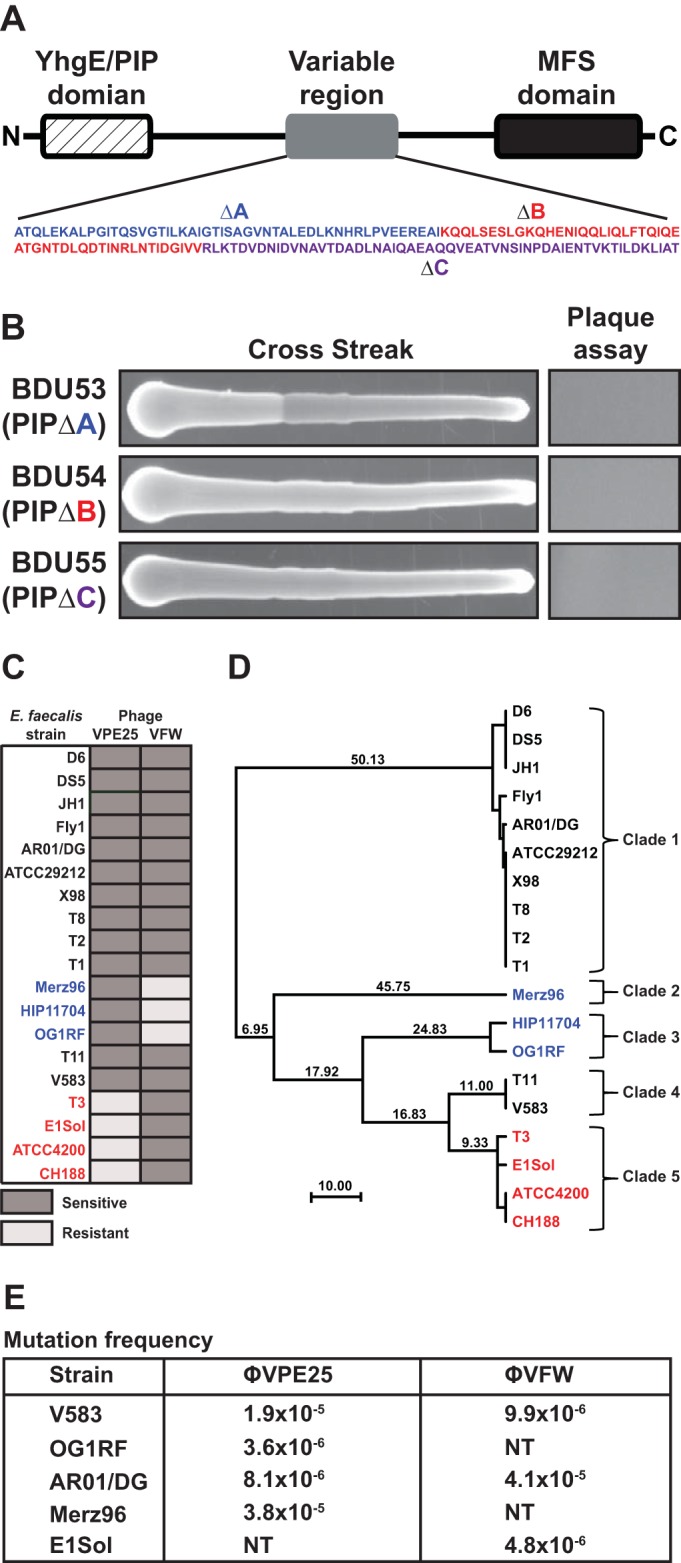
A variable region in PIPEF determines phage tropism in E. faecalis. (A) Schematic of PIPEF from E. faecalis V583. A variable region covering ~160 amino acids is located in the center of the PIPEF coding sequence. (B) Both φVPE25 cross streak and plaque assays show that truncations in the PIPEF variable region abolish phage infectivity regardless of location. The deletions (ΔA, ΔB, and ΔC) correspond to the colored amino acids highlighted in the magnified area of the variable region shown in panel A. (C) Susceptibility profiles of 19 E. faecalis strains for phages φVPE25 and φVFW. (D) Clustering of PIPEF variable region amino acid alignments from 19 strains of E. faecalis. Strains cluster according to their susceptibility patterns as determined in panel C. These strains are indicated by color coding as follows: strains sensitive to killing by both phages (black), strains sensitive to only φVPE25 (blue), and strains sensitive only to φVFW (red). Strains can be further grouped into five specific clades based on PIPEF variable region amino acid identity. (E) Representative clade-specific mutation frequencies for phages φVPE25 and φVFW. NT, not tested (due to natural resistance to the phage of interest).
