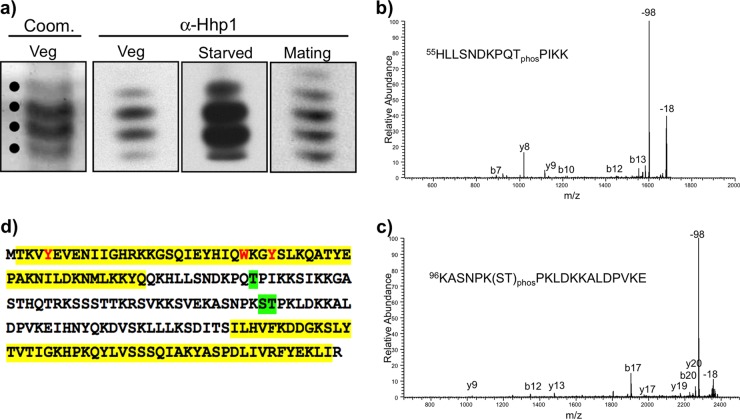
FIG 5 .
Identification of phosphorylation sites on Hhp1. (a) A majority of the Hhp1 isoforms were retained through the isolation procedure. Isoforms were resolved by AU-PAGE after partial purification by reversed-phase HPLC, and a portion of the sample was analyzed by immunoblotting with anti-Hhp1 antiserum. An example of Coomassie-stained bands that were excised for analysis by MALDI-MS/MS (indicated by dots) is shown for vegetatively growing cells (Coom. Veg). (b) Representative MALDI-MS/MS spectrum of an Hhp1 peptide phosphorylated on T64. The signature 98-Da loss for phosphopeptides is indicated, as well as the common 18-Da loss for peptides. Fragment ions are labeled. (c) Representative MALDI-MS/MS spectrum of an Hhp1 peptide phosphorylated at either serine 102 or threonine 103. (d) Amino acid sequence of Hhp1 showing phosphorylation sites that were identified on all Hhp1 isoforms: T64 and S102 or T103 (green highlighting). Yellow highlighting denotes the CD and CSD (the CSD is shortened by 17 amino acids at the N terminus from the original prediction [20] based on recent analysis with HHPRED [59]); methyl-lysine caging residues are red.