Figure 5.
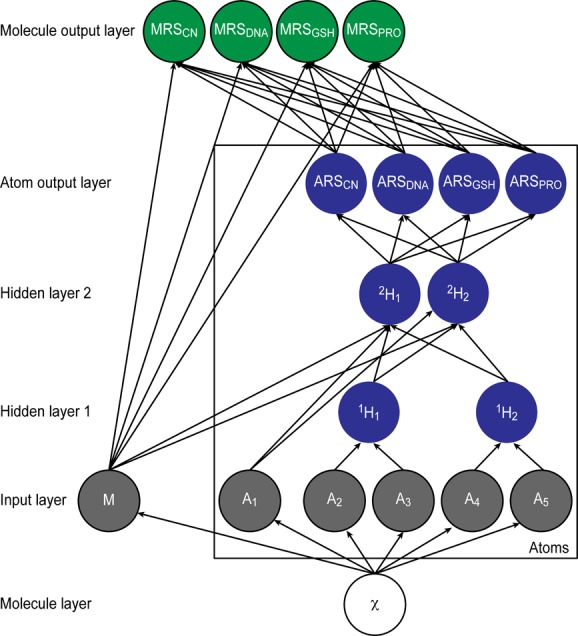
The structure of the XenoSite reactivity model. This diagram illustrates how information flowed through the model, which consisted of one input layer, two hidden layers, and two output layers. By simultaneously modeling all types of reactivity, the model was able to transfer knowledge between related tasks, thereby improving performance substantially over independent models. The model computed atom-level predictions for reactivity to each of four nucleophilic targets: cyanide (ARSCN), DNA (ARSDNA), GSH (ARSGSH), and protein (ARSPRO), collectively referred to as atom reactivity scores (ARS). Additionally, the model computed molecule reactivity scores (MRS): MRSCN, MRSDNA, MRSGSH, and MRSPRO, which predicted the chances of a molecule’s reactivity to each of the four nucleophilic targets, respectively. From the structure of an input model χ, 15 molecule-level and 194 atom-level descriptors were calculated. Some chemically related descriptors, such as neighbor atom identities, were grouped in the first hidden layer (with 30 nodes). Grouped and ungrouped nodes were inputted into the second hidden layer (with 17 nodes), which outputted four atom-level scores. Finally, for each of the four nucleophilic targets, the respective MRS was computed from the top five ARS for each of the four nucleophilic targets, corresponding to the scores of the five atoms predicted within a molecule to be the most reactive to each nucleophile, as well as all molecule-level descriptors. The diagram is condensed and displays one representative molecule input node, five atom input nodes, and two nodes for each hidden layer. The molecule input node is a chemical structure; all other nodes are vectors of real numbers computed from nodes or layers from which there are incoming connections.
