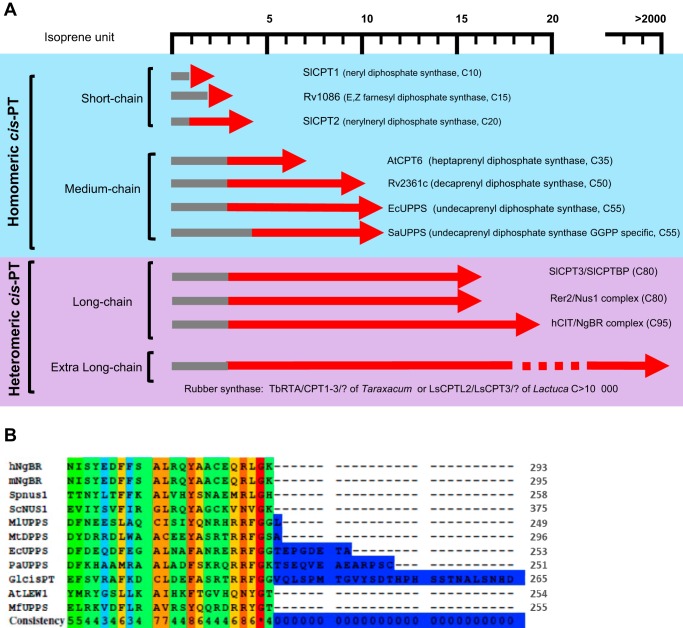
FIGURE 2.
Classification of cis-prenyltransferases. A, classification of cis-PTs according to product chain length and subunit structure. Gray lines indicate numbers of isoprene units of representative allylic substrates for each cis-PT. Red arrows indicate numbers of isoprene units of representative final products. Enzymes represented in the figure are: SlCPT1 (Solanum lycopersicum, GenBankTM NM_001247704), Rv1086 (M. tuberculosis, GenBank: CFB23420.1), SlCPT2 (S. lycopersicum GenBank JX943884), AtCPT6 (A. thaliana, GenBank NP_568882), Rv2361c (M. tuberculosis, GenBank: WP_031739650), EcUPPS (Escherichia coli, GenBank: P60472), SaUPPS (Sulfolobus acidocaldarius, GenBank WP_011277635.1), SlCPT3/SlCPTBP (S. lycopersicum GenBank: JX943885/XP_004241992), hCIT/NgBR (Homo sapiens GenBank accession NP_612468/BAB14439), RER2/NUS1 (S. cerevisiae, GenBank P35196/NP_010088), TbRTA/CPT1–3 (T. brevicorniculatum, GenBank ALX37963/AGE89403/AGE89404/AGE89405), and LsCPTL2/LsCPT3 (L. sativa, GenBank AIQ81190/AIQ81186). B, alignment of the C terminus of NgBR orthologs (hNgBR, human; mNgBR, mouse; SpNus1, S. pombe; ScNus1, S. cerevisiae; AtLEW1, A. thaliana) and single subunit cis-PTs (MlUPPS, Micrococcus luteus; MtDPPS, M. tuberculosis; EcUPPS, E. coli; PaUPPS, P. aeruginosa; GlcisPT, G. lamblia; and MfUPPS, Methanobacterium formicicum). The conservation scoring was performed by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 (*) for the most conserved alignment position.