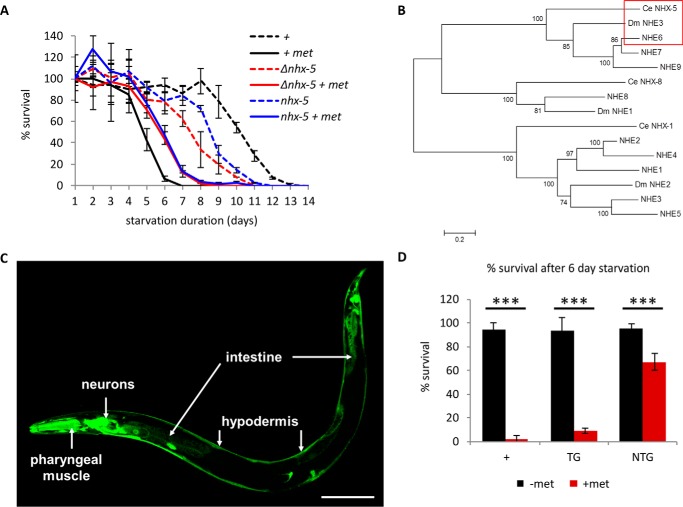
FIGURE 1.
NHX-5 mediates metformin-induced L1 longevity phenotype in C. elegans. A, metformin (100 mm) reduces L1 longevity in wild type, whereas two nhx-5 mutants survive better in the presence of metformin (+ met) (Δnhx-5 and nhx-5). p < 0.001 between metformin-treated wild type and metformin-treated nhx-5 mutant by log-rank test (see “Experimental Procedures”). Each data point is plotted as the mean of triplicated samples and standard deviation. B, phylogenetic tree to compare similarity among endosomal NHEs with C. elegans (Ce) nhx-5 and D. melanogaster (Dm) nhe3. nhe6 and nhe7 are the closest homologs of CeNHX-5 and DmNHE3 (red box). However, NHE6 and CeNHX-5 do not contain the Golgi retention motif, whereas NHE7 does (46). This suggests that NHE6 is the closest functional homolog to ceNHX-5. Five CeNHXs (CeNHX1–5) localize in the plasma membrane and belong to the same phylogenetic group as the human NHE1 family, The phylogenetic tree was created using the MEGA program (47). The p-distance method was used to calculate distance, and the neighbor-joining method was used to create a tree. The tree was evaluated by using the bootstrap method (1000 times). Neighbor-joining, unweighted pair group method with arithmetic mean (UPGMA), and minimum evolution methods created similar trees. C, NHX-5 is expressed in many cells including neurons, pharyngeal muscle, intestine and hypodermis in C. elegans (scale bar = 100 μm). D, transgenic nhx-5 mutant worms carrying a wild-type transgenic copy of nhx-5 (TG for transgenic, black square for without metformin (−met), red square for with metformin (+met) (100 mm)) restore sensitivity to metformin, whereas non-transgenic siblings (NTG, black square for without metformin (−met), red square for with metformin (+met) (100 mm)) are still resistant. Because the difference between wild type and nhx-5 mutants is most obvious after 6 days of starvation (panel A), we compared the percentage of survival between transgenic siblings and non-transgenic siblings after 6 days of starvation. Approximately 120 transgenic worms and 1000 non-transgenic worms were used. The numbers are the mean ± S.D. ***, p < 0.001, by unpaired two-sample t test.