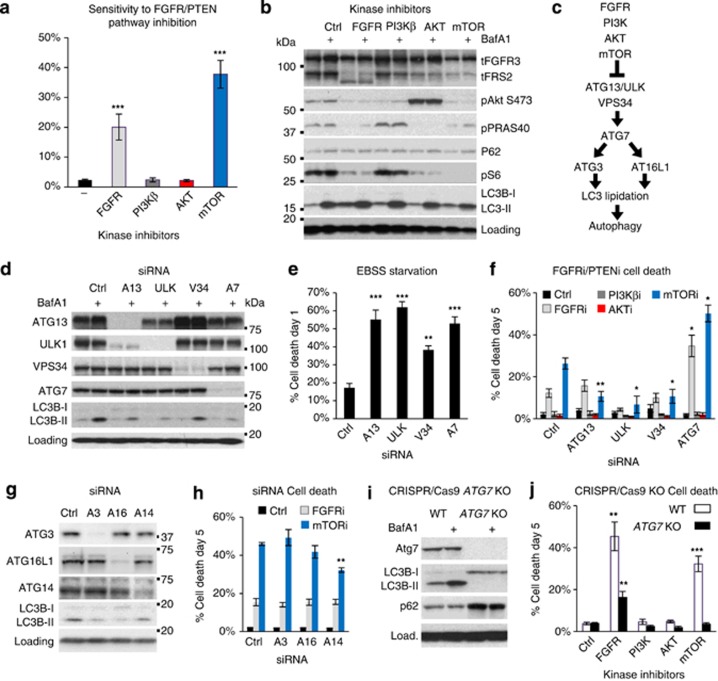
Figure 1.
Activation of autophagy does not contribute to survival under FGFR/PTENi. (a) RT112 cells were treated with the indicated inhibitors at 1 μm for 5 days and assayed for cell death. Histogram shows the number of dead cells (SYTOX Green-positive) as a proportion of total cell number (Hoechst-positive; n⩾7; mean±s.e.). (b) RT112 were incubated with kinase inhibitors for 22 h before the addition of BafA1 (20 nm), where indicated, for the last 2 h. A representative immunoblot confirms target engagement for FGFRi (FRS-2 bandshift), AKTi (hyperphosphorylated in catalytically inactive state), mTORi (S6 dephosphorylation) and BafA1 (LC3B; n=3). (c) Schematic to illustrate mTOR regulation of autophagy. (d–h) RT112 cultures were reverse transfected with the indicated autophagy-targeting siRNAs (10 nm) for 48 h before analysis. (d) Efficiency of protein knockdown and inhibition of autophagic LC3 flux (−/+BafA1) was assayed by immunoblotting at 72 h (n=3). (e and f) Histograms show the quantification of cell death in cultures starved of amino acids and serum (EBSS) for 24 h (e; n=5; mean±s.e.) or treated with the indicated kinase inhibitors for 5 days (n=4; mean±s.e.). (g) Immunoblot confirms knockdown of the indicated autophagy-essential proteins and inhibition of LC3 lipidation. (h) Histogram shows quantification of cell death after treatment with FGFRi or mTORi for 5 days (n=3; mean±s.e.). (i) An ATG7 KO cell line was engineered by CRISPR/Cas9 and immunoblotted to confirm loss of ATG7 expression and LC3 lipidation. (j) ATG7 KO RT112 cultures were assayed for cell death as in (a) (n=4; mean±s.e.). *P<0.05, **P<0.01, ***P<0.001.