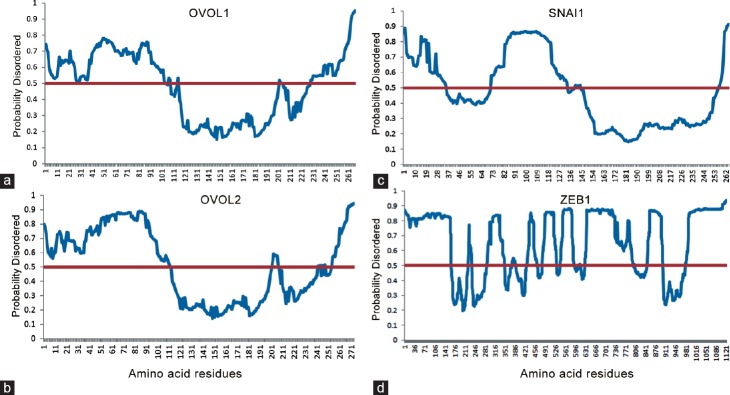
Figure 3.
Disorder prediction for OVOL1/2 (Ovo-Like Zinc Finger 1/2), ZEB1 (Zinc finger E-box-binding homeobox 1), and SNAI1 (Snail Family Zinc Finger 1). The MetaPrDOS metaserver was used to predict the disorder. MetaPrDOS predicts the disorder tendency of each residue using support vector machines from the prediction results of the seven independent predictors. The method has been evaluated by using the Critical Assessment of Techniques for Protein Structure Prediction 7 (CASP7) targets to avoid an overestimation due to the inclusion of proteins used in the training set of some component predictors. As a result, the meta-approach achieves higher prediction accuracy than all methods participating in CASP7. Amino acid numbers are indicated on the X-axis and the disorder likelihood on the Y-axis with a red line indicating the 0.5 (50%) level. (a) OVOL1 (b) OVOL2 (c) SNAI1 (d) ZEB1.