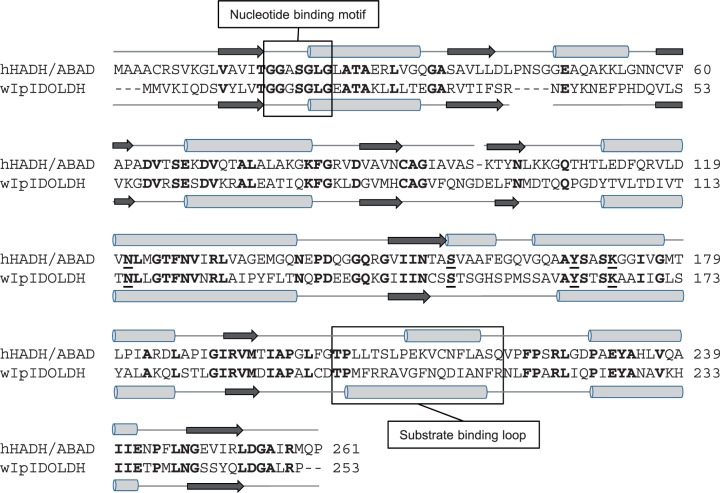
Fig. 4.
Structural comparison of short-chain dehydrogenases. ClustalW alignment of hHADH/ABAD (GenBank NP_004484.1 and western I. pini IDOLDH (wIpIDOLDH; CB408666.1). Conserved residues are in bold font. Gaps inserted to optimize the alignment are indicated by dashes. The substrate binding loops and the nucleotide-binding motifs are boxed. The catalytic tetrad residues are underlined.