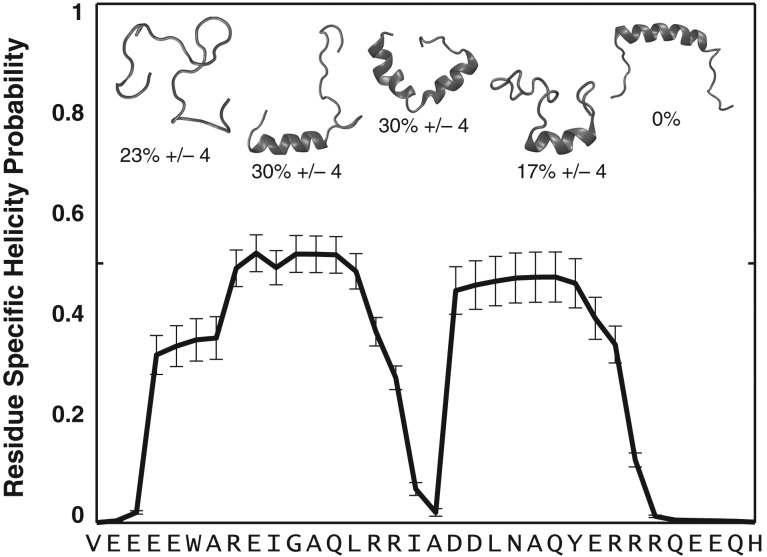
Fig. 2.
The unbound PUMA adopts a heterogeneous conformational ensemble. The figure summarizes results from all atom ABSINTH-based simulations of PUMA. The sequence prefers a heterogeneous ensemble of conformations. These include conformations with independent N- and C-terminal helical halves, coil-like N- or C-terminal halves that are populated with helical C- or N-terminal halves, and fully coil-like conformations. The heterogeneity is quantified in terms of the percent probabilities associated with distinct conformational types. These populations are used to quantify a residue-specific helicity profile that quantifies the percent probability of finding a residue as part of a regular alpha helical segment that is at least six residues long. Note that in the simulations the central helix conformation is not accessed by the wild type sequence of PUMA.