Abstract
Aberrant mitochondrial fission plays a pivotal role in the pathogenesis of skeletal muscle insulin resistance. However, fusion-fission dynamics are physiologically regulated by inherent tissue-specific and nutrient-sensitive processes that may have distinct or even opposing effects with respect to insulin sensitivity. Based on a combination of mouse population genetics and functional in vitro assays, we describe here a regulatory circuit in which peroxisome proliferator–activated receptor γ (PPARγ), the adipocyte master regulator and receptor for the thiazolidinedione class of antidiabetic drugs, controls mitochondrial network fragmentation through transcriptional induction of Bnip3. Short hairpin RNA–mediated knockdown of Bnip3 in cultured adipocytes shifts the balance toward mitochondrial elongation, leading to compromised respiratory capacity, heightened fatty acid β-oxidation-associated mitochondrial reactive oxygen species generation, insulin resistance, and reduced triacylglycerol storage. Notably, the selective fission/Drp1 inhibitor Mdivi-1 mimics the effects of Bnip3 knockdown on adipose mitochondrial bioenergetics and glucose disposal. We further show that Bnip3 is reciprocally regulated in white and brown fat depots of diet-induced obesity and leptin-deficient ob/ob mouse models. Finally, Bnip3−/− mice trade reduced adiposity for increased liver steatosis and develop aggravated systemic insulin resistance in response to high-fat feeding. Together, our data outline Bnip3 as a key effector of PPARγ-mediated adipose mitochondrial network fragmentation, improving insulin sensitivity and limiting oxidative stress.
Introduction
Whereas the systemic metabolic benefit ascribed to regulated mitochondrial energy dissipation in beige and brown fat depots is now widely appreciated (1), the physiological relevance of white adipose tissue (WAT) mitochondria has remained enigmatic. By coordinating key biochemical processes central to WAT, ranging from lipogenesis, glyceroneogenesis, and adipokine biology, white fat mitochondria may play a critical role in determining systemic insulin sensitivity (2). Notably, multiple studies have shown that mitochondrial biogenesis, fusion-fission dynamics, bioenergetics, and transcript levels of nuclear-encoded mitochondrial genes are globally disrupted in visceral/gonadal WAT (gWAT) of both dietary and genetic murine obesity models (3–6). Of interest, the thiazolidinedione (TZD) class of antidiabetic drugs induces dramatic changes in adipocyte mitochondrial bioenergetics, at least in part by ameliorating the obesity-related impairment of mitochondrial gene expression (3–5). TZDs are known ligands for the nuclear hormone receptor peroxisome proliferator–activated receptor γ (PPARγ), the master regulator of fat cell biology (7). In agreement, studies using in vitro models of adipogenesis have unveiled that mitochondrial biogenesis and network remodeling are integral to the transcriptional program controlled by PPARγ (8–11). This raises the prospect that correcting mitochondrial (dys)function in gWAT may contribute to the insulin-sensitizing action of TZDs.
Mitochondrial fusion, fission, and selective mitochondrial autophagy (mitophagy) form an essential axis of quality control (QC). This integrated network of cellular pathways, collectively referred to as mitochondrial dynamics, serves to limit oxidative damage caused by reactive oxygen species (ROS) generated as side-product of the respiratory chain (mitochondrial ROS [mROS]). Dysregulated OXPHOS and mROS generation are intimately linked to the development of obesity-related insulin resistance (IR). Of note, chronic nutrient overload has recently been suggested to interfere with fusion-fission dynamics, leading to progressive mitochondrial dysfunction (12). In agreement, disruption of mitochondrial fusion is instrumental to the pathogenesis of skeletal muscle IR in both mice and humans (13–15). Paradoxically, a regulated shift from fused, elongated mitochondrial networks to fragmented, punctate structures in adipocytes is a hallmark of ligand activation of PPARγ (9–11); this shift most likely occurs via transcriptional induction of hitherto unknown target genes that impose on the fusion-fission apparatus. This indicates that mitochondrial dynamics are regulated by tissue-specific and nutrient-sensitive processes that may variably affect insulin sensitivity. Thus research aimed at identifying PPARγ effectors that control the fusion-fission balance in gWAT and their respective roles in modulating insulin sensitivity may yield novel insights into the antidiabetic effects of PPARγ agonism.
As part of a bioinformatics approach aimed to identify potential regulators of adipose mitochondrial fusion-fission balance, BCL2/adenovirus E1B 19-kDa interacting protein 3 (Bnip3) emerged as a promising candidate. Bnip3 and the functionally related Nix protein comprise a conserved subfamily of atypical BH3-only members. Bnip3 and Nix form stable homodimeric complexes via a unique COOH terminal transmembrane domain (TMD) that is required for mitochondrial targeting (16). Of interest, Bnip3 regulates mitochondrial dynamics by interacting with the core fusion-fission machinery (17,18). In addition, Bnip3 and Nix are potent inducers of autophagy and mitophagy through shared domain-specific activities. The proautophagic activity has been ascribed to their BH3 domain, which disrupts the mitochondrial BCL2–BECN1 complex, leading to BECN1-dependent autophagosome synthesis (19,20). By means of an LC3-II-interacting region (LIR) facing the cytosol, Bnip3 and Nix serve as mitophagy receptors by tethering mitochondria to LC3-II present on nascent autophagosomes (21–23).
In this article we describe a PPARγ-Bnip3 axis that couples mitochondrial network fragmentation to adipose insulin sensitization. In addition, we provide evidence in support of fusion-fission regulation as a novel mechanism to limit fatty acid (FA)–induced mROS generation and oxidative damage. Finally, Bnip3−/− mice develop systemic metabolic dysfunction in response to high-fat (HF) feeding.
Research Design and Methods
In Silico Search
Genome-wide mRNA profiling was performed using GeneNetwork (24) (http://www.genenetwork.org) on an ApoE null F2 intercross between C57BL/6J X C3H/HeJ (BHF2) mice (25). The correlation coefficients between systemic phenotypes and adipose gene expression were transformed into a heatmap. Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways with overrepresentation in the Bnip3 set were defined using ClueGo (26). PPARγ chromatin immunoprecipitation sequencing (GSE13511) was performed using HOMER (27).
RNA/DNA Isolation and RT-PCR
RNA isolation and RT-PCR analysis were performed as described previously (28). mRNA expression was calculated by the ΔΔCt method and normalized to 36B4. Total DNA was isolated with the DNeasy kit (Qiagen), and mitochondrial DNA (Cox2, CytB) was normalized to nuclear DNA (Glucagon, β-Globin). Primer sequences are available upon request.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl [pH 7.4], 2 mmol/L EDTA [pH 8.0], 10 mmol/L NaF, 1 mmol/L Na3VO4, 0.5% sodium deoxycholate, 1 mmol/L phenylmethylsulfonyl fluoride, and 1% Triton X-100) containing protease inhibitors. Immunoblotting was performed as described elsewhere (28). Antibodies, manufacturers, and dilutions are listed in Supplementary Table 1.
Plasmids and Lentiviral Expression Constructs
Lentiviral stocks were generated as described elsewhere (28). pLKO.1-puro SHC004 served as the internal control in our studies. pLKO.1-puro or pLKO_TRC005 short hairpin RNAs (shRNAs) targeting Bnip3 were the following: shB3_1 (no. 229457), shB3_2 (no. 9691), shB3_3 (no. 9689), and shB3_4 (no. 9687). Full-length DNA sequences of Bnip3 and Nix were amplified by PCR and cloned into gateway entry plasmid pDONR221 (Invitrogen). Bnip3 ∆TMD or ∆LIR (W13A/L16A) were generated by site-directed mutagenesis (Stratagene). Inducible expression constructs were generated by attL × attR recombination between pDONR221 and pInducer-Bla, genetically modified from the pINDUCER20 lentiviral vector (a gift from Stephen Elledge [29]; Addgene plasmid no. 44012), by replacing a neomycin cassette with the blasticidin resistance gene.
Cell Culture
All cell lines were maintained in DMEM/10% FBS supplemented with antibiotics. Polyclonal 3T3-L1 preadipocyte cell lines stably expressing shRNAs were generated by lentiviral transduction and selection with puromycin. Where indicated, 3T3-L1 preadipocytes were also transduced with pInducer-Bla and selected with blasticidin. Differentiation was initiated 3 days after confluence by exposure to 0.5 mmol/L isobutylmethylxanthine (IBMX), 1 µmol/L dexamethasone, 0.59 µg/mL insulin, and 1 µmol/L rosiglitazone for 48 h, followed by 0.59 µg/mL insulin for 5 days. Where indicated, doxycycline (1 µg/mL) was added to cell culture media from day 4 onward. Primary mouse embryo fibroblasts were differentiated under the same conditions, but with 1.5 µg/mL insulin. T37i cells (a gift from Esther Lutgens) were differentiated with 2 nmol/L triiodothyronine and 0.59 µg/mL insulin.
2-Deoxyglucose and 3-Methylglucose Uptake
3T3-L1 adipocytes were incubated in HEPES buffer (20 mmol/L HEPES, 1.3 mmol/L CaCl2, 132 mmol/L NaCl, 1 mmol/L MgSO4, 1.2 mmol/L KH2PO4, 6 mmol/L KCl; pH 7.4) containing 1.5 mmol/L pyruvate and 0.2% BSA and stimulated with insulin for 15 min. 2-Deoxyglucose (2-DG) uptake was initiated by adding 0.2 µCi/ml 2-[14C]-deoxyglucose radiolabel (Perkin Elmer) to 100 µmol/L for 10 min. 3-O-[14C]-methylglucose (3-MG) uptake was performed as described elsewhere (30). Radiolabel uptake was normalized for protein and nonspecific diffusion (5 µmol/L cytochalasin B).
Flow Cytometric Analysis of Mitochondrial Metabolism and GLUT4 Translocation
Cells were incubated in HEPES buffer (containing 5 mmol/L glucose) for 30 min. For analysis of ∆ψm, 3T3-L1 adipocytes were incubated with 2.5 µmol/L JC-1 (30 min). Cells were harvested in ice-cold PBS (5 mmol/L EDTA, 0.2% BSA) and analyzed by FACS. mROS production was analyzed with either MitoSOX (2 µmol/L; 45 min) or dihydroethidium (DHE) (2 µmol/L; 30 min). Analysis of 7XMyc-GLUT4-GFP exocytosis was performed as described previously (31).
Immunofluorescence Microscopy
Differentiated 3T3-L1 adipocytes were reseeded to confluence on glass coverslips and cultured in DMEM with 0.59 µg/mL insulin for 48 h and fixed in 4% paraformaldehyde/PBS (5% sucrose) for 15 min. After permeabilization (PBS, 0.2% Triton-X100, 1% BSA) for 20 min, quenching (PBS, 0.2 M glycine, 3% BSA) for 30 min, and saturation (PBS, 3% BSA) for 2 h, coverslips were probed with rabbit anti-Tom20 (PBS, 2% BSA, 0.02% Tween-20) overnight at 4°C. Coverslips were washed, incubated with Alexa-568-conjugated goat-antirabbit IgG for 1 h at room temperature, mounted in DAPI, and analyzed by epifluorescence microscopy (DM5500; Leica, Germany) with a HCX-PL-APO 63×/1.40–0.60 oil-immersion objective. Mitochondrial morphology was quantified essentially as previously described (15).
Bioenergetics Profile
Cellular respiration was determined with a Seahorse XF96 Extracellular Flux Analyzer. 3T3-L1 adipocytes were reseeded to 25,000 cells/well, cultured overnight in DMEM (0.59 µg/mL insulin), and then cultured for another 16 h in maintenance medium. Thereafter, cells were incubated in XF Base medium (5 mmol/L glucose, 0.2% BSA). The bioenergetics profile was analyzed by sequential injections of oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), and rotenone/antimycin.
Animals and Diets
C57BL/6 and ob/ob mice were from Jackson Laboratories, and Bnip3−/− mice were generated as described previously (32). Mice were housed in a climate-controlled environment with a 12-h light/12-h dark cycle and ad libitum access to water and food. Wild-type (WT) and ob/ob mice received a chow diet (AM-II) for 12 weeks before analysis. For diet-induced obesity, 8-week-old male WT and Bnip3−/− mice were fed either a HF (60% kcal fat; D12492; Research Diets Inc.) or low-fat (LF) diet (10% kcal fat; D12450B) for 16 weeks. Tissues were collected and snap-frozen in liquid nitrogen or fixed in 4% formalin. Animal care and study protocols were approved by the Ethical Committee for Animal Experiments and were in accordance with Dutch law.
Metabolic Phenotyping of Bnip3−/− Mice
Blood glucose concentrations were determined with a glucometer. Plasma insulin (Crystal Chem Inc.) and leptin concentrations (EMD Millipore) were measured by ELISA. Plasma triglycerides (TGs) and free FAs (FFAs) were measured using kits from Biolabo and Wako Chemical, respectively. An oral glucose tolerance test was performed by oral gavage of glucose (2 g/kg body weight [BW]) after 4 h of fasting. Blood glucose concentrations were analyzed by tail vein puncture before and 15–120 min after glucose loading. Metabolites were determined using blood (week 12) and plasma (week 16), as described elsewhere (33).
Immunohistochemistry and Adipocyte Size
Tissues were fixed in 4% formalin and embedded in paraffin. Deparaffinized tissue sections (4 µmol/L) were stained with hematoxylin-eosin. Immunohistochemistry was performed with rabbit anti-BNIP3, followed by alkaline phosphatase–conjugated goat antirabbit IgG. Bound alkaline phosphatase activity was detected using Vector Blue substrate (Vector Laboratories, Burlingame, CA) in the presence of levamisole (1 mmol/L). gWAT cell size was determined by acquiring images from nonoverlapping fields, followed by Adiposoft analysis. The average area and diameter of >200 cells were determined and expressed in arbitrary units.
Statistics
Values are presented as mean ± SEM. Statistical significance was assessed with a two-tailed Student t test (unpaired). The criterion for statistical significance was set at P < 0.05.
Results
Mouse Population Genetics Links Adipose Mitochondrial Dynamics to Markers of Insulin Sensitivity
F2 intercrosses of inbred mouse strains yield genotypically distinct offspring, randomly segregating for their innate disease susceptibilities. Here we studied an apolipoprotein E null F2 intercross of C57BL/6J × C3H/HeJ inbred mice, enabling robust analysis of adipocyte microarray data coupled to systemic metabolic phenotypes after HF feeding. This poses a powerful model to infer the physiological relevance of WAT mitochondrial dynamics.
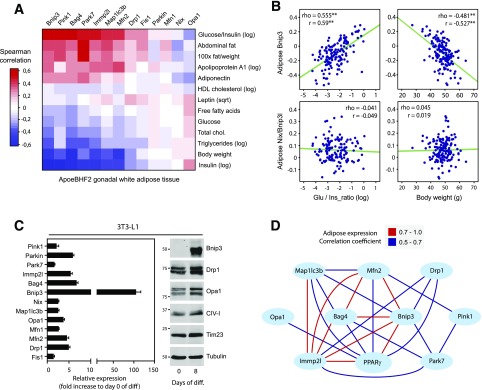
Specifically, we screened whether genetic variation in mRNA levels of known mitochondrial dynamics and QC-related genes in gWAT correlated with systemic metabolic phenotypes. Importantly, mRNA expression selectively enriched for the phosphatase and tensin homolog–induced putative kinase 1 (Pink1)–Parkin axis showed highly significant correlations with measures of insulin sensitivity (Fig. 1A). Pink1 and Parkin are integral to a variety of QC processes, including mitophagy, fusion-fission balance, biogenesis, and repair (34). In addition, BH3-only Bnip3 was one of the top-ranked genes whose expression positively correlated with the glucose-to-insulin ratio (Fig. 1A). Conversely, transcript levels of the functionally related Nix showed virtually no correlations to systemic parameters (Fig. 1A). Individual correlation plots for adipose Bnip3 and Nix gene expression to the glucose-to-insulin ratio and BW are illustrated in Fig. 1B. QC-related mRNA levels also inversely correlated with BW, plasma insulin concentrations, and lipid concentrations. This was accompanied by a positive covariation with measures for adiposity.
Figure 1.
Identification of Bnip3 as a potential PPARγ effector regulating adipose mitochondrial dynamics. A: Heatmap transformation of gene/trait correlations for mitochondrial dynamics–related mRNA transcripts in gWAT versus the indicated metabolic phenotypes scored across individual males from the BxH apolipoprotein E null F2 mouse cross (BHF2; n = ∼140). The axes were hierarchically clustered (Euclidean distance). B: Correlation matrix plots between adipose Bnip3 and Nix transcript levels and phenotypic traits, glucose-to-insulin ratio, and BW (r = Pearson and rho = Spearman). C: Quantitative PCR and immunoblot analyses of mitochondrial dynamics–related expression in 3T3-L1 adipocytes (n = 4). D: Pparγ mRNA expression shows covariance with mitochondrial dynamics–related transcript levels in gWAT. Red lines indicate a positive Pearson correlation coefficient of 0.7–1.0, and blue lines signify a coefficient of 0.5–0.7. Phenotypic trait nomenclatures are as follows: abdominal fat (grams), cholesterol (milligrams per deciliter), FFAs (milligrams per deciliter), HDL fraction of cholesterol log (milligrams per deciliter), glucose (milligrams per deciliter), plasma insulin log (picograms per liter), TGs log (milligrams per deciliter). diff, differentiation; glu, glucose; ins, insulin; log, log transformed; sqrt, square root transformed. **P < 0.01.
The systemic metabolic benefit associated with high QC mRNA levels in gWAT led us to explore whether these genes were regulated as part of the PPARγ-driven transcriptional program. Notably, gene expression analysis in the 3T3-L1 in vitro model for adipogenesis revealed a robust transcriptional induction of Bnip3 (Fig. 1C). We next addressed the Bnip3-associated transcriptional network in mouse F2 populations. Bnip3 mRNA expression in gWAT had strong covariance with genes related to intermediary metabolism and PPARγ signaling (Supplementary Fig. 1A). Importantly, we observed significant coregulated expression among fusion-fission and mitophagy-related genes with PPARγ (Fig. 1D). These findings indicate the physiological relevance of adipocyte mitochondrial dynamics and define Bnip3 as a promising candidate for the PPARγ effects on the fusion-fission balance.
Bnip3 Expression Is Regulated During Adipocyte Differentiation
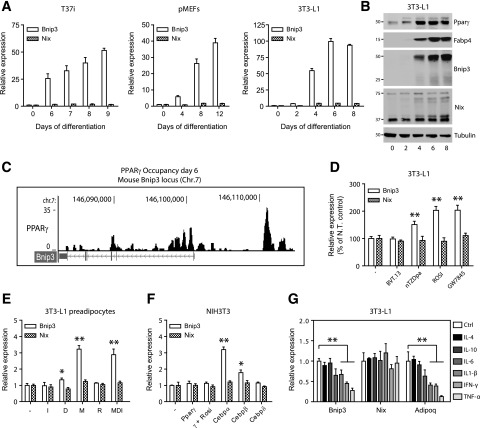
To validate Bnip3 as a de novo adipogenic target gene, we analyzed a time course in several in vitro models for white and brown adipocyte differentiation. This revealed a universal increase in Bnip3 mRNA, whereas Nix was constitutively expressed (Fig. 2A). Accordingly, Bnip3 protein levels were robustly detected in the course of adipogenesis, at a time consistent with its regulation by adipose-specific transcription factors such as PPARγ (Fig. 2B). Through analysis of global PPARγ DNA-binding data (27), we found several putative PPARγ binding regions in the mouse Bnip3 locus (Fig. 2C). In agreement, Bnip3 mRNA was hyperinduced in 3T3-L1 adipocytes upon incubation with synthetic PPARγ ligands (Fig. 2D).
Figure 2.
Bnip3 is highly regulated during adipocyte differentiation. A: Relative Bnip3 and Nix mRNA levels during a time course of differentiation in T37i hibernoma cells (left panel), primary mouse embryo fibroblasts (pMEFs; middle panel), or 3T3-L1 cells (right panel). B: Bnip3 and Nix protein levels in 3T3-L1 preadipocytes exposed to MDI. Note that Bnip3 protein is almost exclusively present in functionally “active” homodimers (50 kDa), whereas Nix is constitutively expressed as the “inactive” monomeric form (37 kDa). PPARγ and Fabp4 were included as positive controls of the adipogenic process. C: Chromatin immunoprecipitation sequencing profile of PPARγ binding sites within the mouse Bnip3 locus from 3T3-L1 cells differentiated with MDI for 6 days. These data are from the sequencing study by Nielsen et al. (27). D: Relative Bnip3 and Nix mRNA levels in 3T3-L1 adipocytes differentiated for 8 days and treated with the indicated PPARγ ligands (1 µmol/L) for another 24 h. N.T., nontreated. E: Relative Bnip3 and Nix mRNA levels in confluent 3T3-L1 preadipocytes treated with individual components of the MDI cocktail for 48 h. D, 1 µmol/L dexamethasone; M, 0.5 mmol/L IBMX; I, 0.59 µg/mL insulin; R, 1 µmol/L rosiglitazone (Rosi). F: Relative Bnip3 and Nix mRNA levels in the fibroblastic NIH3T3 cell line after transfection of the indicated plasmids encoding adipogenic transcription factors for 36 h. G: Relative Bnip3 and Nix mRNA levels in 3T3-L1 adipocytes differentiated for 8 days and exposed to the indicated pro- and anti-inflammatory cytokines for 24 h (50 ng/mL each). Values are expressed as mean ± SEM (n = 3 or 4 in A and D–G). *P < 0.05; **P < 0.01.
We next evaluated which component of the adipogenic cocktail regulated Bnip3 expression. We stimulated confluent 3T3-L1 preadipocytes with IBMX (M), dexamethasone (D), insulin (I), or the PPARγ ligand rosiglitazone. Only IBMX or the combination of MDI led to enhanced Bnip3 transcription, indicating that Bnip3 is primarily responsive to changes in cellular cAMP levels (Fig. 2E). The ability of cAMP signals to activate the CCAAT/enhancer binding protein (C/EBP) family of transcription factors prompted us to test whether C/EBPs regulate Bnip3 expression. Transfection assays with plasmids encoding adipogenic regulators into NIH3T3 cells revealed enhanced Bnip3 transcription by C/EBPα and to lesser extent by C/EBPβ (Fig. 2F). These data suggest that Bnip3 is regulated by the concerted actions of C/EBPα and PPARγ. Finally, Bnip3 expression was similarly mitigated as known adipogenic genes when subjected to a panel of proinflammatory cytokines associated with obesity-induced gWAT inflammation (Fig. 2G).
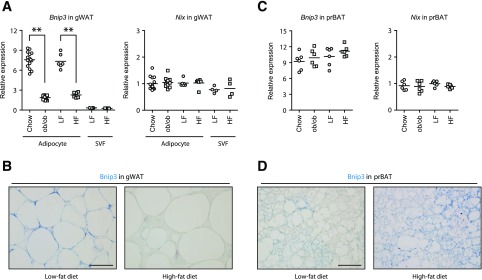
To confirm our in vitro results, we analyzed Bnip3 expression in white and brown fat depots in murine models of dietary and genetic obesity. Whereas Bnip3 and Nix homologs were both expressed in gWAT of lean WT controls, only Bnip3 was strikingly downregulated in HF diet–induced obesity and leptin-deficient ob/ob mice (Fig. 3A and B and Supplementary Fig. 1B). Subfractionation of gWAT validated that Bnip3 is exclusively expressed in adipocytes and not in the stromal vascular fraction (Fig. 3A). In addition, mitochondrial dynamics–related transcript levels in gWAT were similarly downregulated in murine models of obesity (Supplementary Fig. 1C). Conversely, Bnip3 mRNA levels in brown fat were unaffected by chronic overnutrition (Fig. 3C). Intriguingly, Bnip3 protein levels were actually elevated in response to HF feeding (Fig. 3D and Supplementary Fig. 1D), indicating posttranslational regulation. In aggregate, these data strongly suggest that adipose Bnip3 expression is regulated in a depot-specific and nutrient-sensitive fashion.
Figure 3.
Bnip3 is reciprocally regulated in white and brown fat depots of murine obesity models. A: Relative Bnip3 and Nix mRNA levels in gWAT of WT mice and ob/ob mice fed a chow diet for 12 weeks (n = 12–15) or in gWAT (n = 6) and the stromal vascular fraction (SVF) (n = 4) of WT mice fed an LF or HF diet for 16 weeks. B: Immunohistochemical staining for Bnip3 in gWAT from mice fed LF and HF diets. C: Relative Bnip3 and Nix mRNA levels in perirenal BAT (prBAT) of WT mice and ob/ob mice fed a chow diet for 12 weeks (n = 6) or WT mice fed an LF or HF diet (n = 6). D: Immunohistochemical staining for Bnip3 in prBAT from mice fed LF and HF diets. Scale bars, 50 μm. **P < 0.01.
Bnip3 Regulates Adipose Mitochondrial Fusion-Fission Dynamics and Bioenergetics
Bnip3 promotes mitochondrial fission when ectopically expressed or in the context of hypoxia (17,18,35). To examine whether Bnip3 is a bona fide PPARγ effector that controls fusion-fission balance, we used lentiviral shRNAs to generate stable 3T3-L1 cell lines (shB3_1–3) with effective Bnip3 knockdown (KD) (Fig. 4A and B). Bnip3-KD cells exhibited reduced differentiation capacity after adipogenic induction with the standard MDI cocktail (Supplementary Fig. 2A and B), as reported previously for the depletion of core fission-related genes (10). To circumvent potentially confounding effects of reduced adipogenesis on our analysis, we opted to include rosiglitazone during the initial phase to boost differentiation, which largely corrected adipogenic parameters in Bnip3-KD cells (Supplementary Fig. 2C and D).
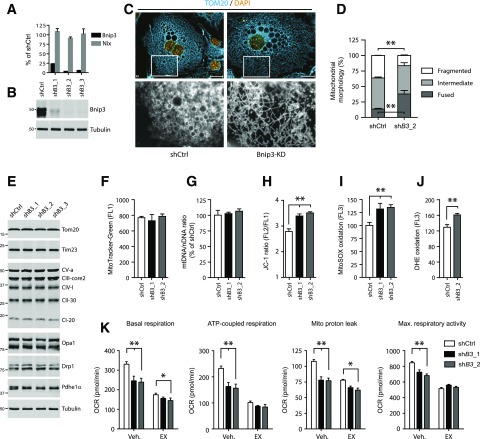
Figure 4.
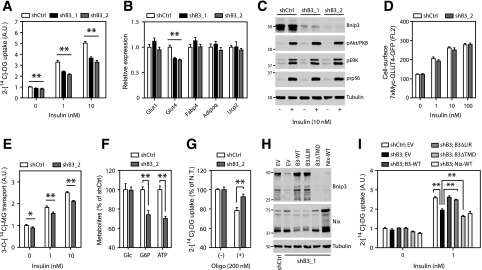
Adipose Bnip3 modulates mitochondrial fusion-fission balance and bioenergetics. A: Relative Bnip3 and Nix mRNA levels (expressed as a percentage of shCtrl). B: Immunoblot analysis of Bnip3 protein in 3T3-L1 adipocytes stably transduced with shCtrl or shRNAs targeting Bnip3 (shB3_1–3) and differentiated for 7 days. C and D: Representative image of (C) (bottom row, magnification ×10) and quantified mitochondrial morphology (D) in shCtrl or shB3_2 adipocytes and immunostained for endogenous Tom20 protein. Morphology was scored as follows: fragmented, punctuate and round; intermediate, mixture of round and shorter tubulated; elongated, long and high interconnectivity. Values are expressed as a percentage of the total number of cells counted (≥100 per experiment). E: Immunoblot analysis of multiple markers for mitochondrial function and metabolism. F: Flow cytometric analysis of MitoTracker Green staining. G: The mitochondrial-to-nuclear DNA ratio. Flow cytometric analysis of the ∆ψm using JC-1 (H) and mROS formation using MitoSOX (I) and DHE (J). K: Mitochondrial bioenergetics profile. Basal OCR, ATP-linked and proton leak–associated respiration (oligomycin; 2 µmol/L) and maximal respiratory capacity (FCCP; 0.75 µmol/L) were examined using a Seahorse XF96 extracellular flux analyzer. Where indicated, the CPT1-inhibitor EX (100 µmol/L) or H2O vehicle (Veh.) was injected during the assay. Max., maximum. Values are expressed as mean ± SEM (n = 3 in A, C, and D; n = 4 in F–J; and n = 7 or 8 in K). Scale bars, 20 μm. *P < 0.05; **P < 0.01.
Subcellular fractionation substantiated mitochondrial targeting of adipose Bnip3 (Supplementary Fig. 2E). We next addressed fusion-fission status by immunostaining for the mitochondrial protein Tom20. Of note, Bnip3 KD led to a robust increase in the percentage of cells with fused, elongated mitochondrial networks (38% vs. 13.5%) relative to cells expressing a control shRNA (shCtrl) (Fig. 4C and D). Accordingly, we observed a lower percentage of cells with fragmented mitochondria (16% vs. 36%) upon Bnip3 KD, supporting our contention that Bnip3 stimulates fission in adipocytes. Electron microscopy analysis showed smaller and less electron-dense mitochondria in shCtrl adipocytes (Supplementary Fig. 2F).
To address the functional relevance of altered fusion-fission dynamics, we studied related processes such as QC and bioenergetics. Bnip3 is known to induce mitophagy and/or autophagy in a variety of cell lines (19–22). However, we found no evidence of altered mitochondrial content and turnover upon Bnip3 KD in 3T3-L1 adipocytes (Fig. 4E–G; Supplementary Fig. 2G). In addition, autophagic flux was similar in shCtrl and Bnip3-KD cells, as monitored by LC3-II appearance in the presence of the v-ATPase inhibitor bafilomycin A1 (Supplementary Fig. 2H), suggesting that Bnip3 is dispensable for mitophagy.
To study the effects of Bnip3 inhibition on mitochondrial bioenergetics, we first loaded 3T3-L1 adipocytes with JC-1 to assess the mitochondrial membrane potential (∆ψm). Flow cytometric analysis of JC-1 stained cells revealed that Bnip3 KD augmented the ∆ψm (Fig. 4H). This was accompanied by heightened radical formation, as reflected by enhanced oxidation of the ROS probes MitoSOX and DHE (Fig. 4I and J). In agreement, protein carbonylation analyses in 3T3-L1 lysates showed an increase in oxidative damage upon Bnip3 KD (Supplementary Fig. 2I). Of note, despite similar markers for mitochondrial mass (Fig. 4E–G), shCtrl cells showed a higher oxygen consumption rate (OCR), both at baseline and under FCCP-induced maximal respiratory conditions, relative to Bnip3 KD (Fig. 4K). A significant portion of the higher baseline OCR was oligomycin-sensitive, indicating increased ATP-linked respiration in shCtrl adipocytes. The oligomycin-insensitive OCR itself (caused by a proton leak) was also slightly elevated compared with Bnip3 KD (Fig. 4K). Hence adipose induction of Bnip3 increases both energetic demand and the degree of uncoupled respiration.
Since respiration with FAs as substrates is typically associated with mitochondrial network fragmentation, inherent proton leak, and lower values of ∆ψm (12), we reasoned that the Bnip3-regulated increase in OCR could be the result of an enhanced rate of FA β-oxidation (FAO). Indeed, acute exposure to etomoxir (EX), an irreversible CPT1 inhibitor and FA import into mitochondria, lowered OCR indices of shCtrl adipocytes to Bnip3-KD levels (Fig. 4K). Collectively, these results suggest that dysregulated mitochondrial dynamics is causal for diminished respiration and elevated oxidative stress in Bnip3-KD adipocytes.
Bnip3 KD Impairs Insulin-Stimulated Glucose Uptake in 3T3-L1 Adipocytes
PPARγ-induced changes in white adipocyte mitochondrion-related gene expression correlate significantly with insulin sensitivity in animal models, as shown here (Fig. 1) and by others (3,6). We therefore probed Bnip3-KD adipocytes for their capacity to facilitate glucose transport. Importantly, insulin-stimulated 2-DG uptake was significantly reduced by ∼33% and 40% in shB3_1 and shB3_2 cell lines, respectively, relative to shCtrl (Fig. 5A). This correlated in part with decreased Glut4 transcript levels in Bnip3-KD adipocytes (Fig. 5B). Conversely, insulin-stimulated phosphoinositide 3-kinase/Akt signaling and GLUT4 exocytosis were both unaffected by Bnip3 KD (Fig. 5C and D).
Figure 5.
Bnip3 KD impairs insulin-stimulated glucose uptake in 3T3-L1 adipocytes. A: Basal and insulin-mediated 2-DG uptake in 3T3-L1 adipocytes transduced with either shCtrl, shB3_1 or shB3_2 and differentiated for 8 days. B: Relative mRNA levels of adipogenic markers. C: Phospho-Akt (p-Akt)/PKB, phospo-ERK1/2 (pERK), and phospho-rpS6 (prpS6) protein levels in 3T3-L1 adipocytes starved for 2 h and treated with or without insulin (10 nmol/L) for 15 min. D: Flow cytometric analysis of 7XMyc-GLUT4 plasma membrane occupancy in 3T3-L1 adipocytes transduced with shCtrl or shB3_2, together with 7XMyc-GLUT4-GFP, starved for 5 h, and incubated with insulin (0–100 nmol/L) for 15 min. E: Basal and insulin-stimulated 3-MG transport. A.U., arbitrary units. F: Analysis of steady-state levels of cellular ATP, glucose (Glc), and glucose-6-phosphate (G6P). G: Insulin-stimulated 2-DG uptake in the presence of oligomycin (200 nmol/L) for 2 h. Nontreated (N.T.) values are expressed as 100% for both shCtrl and shB3_2 to address the relative sensitivity to oligomycin. H and I: Bnip3 and Nix protein levels (H) and basal and insulin-stimulated 2-DG transport (I) in shCtrl or in shB3_1 adipocytes stably transduced with inducible constructs expressing the indicated Bnip3 deletion mutants. Values are expressed as mean ± SEM (n = 3 or 4 in A–I, except for n = 6 in D and F). *P < 0.05; **P < 0.01.
Impaired 2-DG uptake may reflect a lesion in glucose transport across the plasma membrane or in retention via hexokinase-driven phosphorylation. Therefore we also determined uptake of the nonphosphorylatable analog 3-MG in response to insulin, which specifically mirrors the rate of transport across GLUT4. Notably, the inhibitory effect of Bnip3 KD on insulin-induced glucose disposal in 3T3-L1 adipocytes was less pronounced with 3-MG as the glucose analog than with 2-DG (Fig. 5E), suggesting that Bnip3 exerts control over the rate of glucose phosphorylation. In agreement, we detected increased steady-state levels of ATP and glucose-6-phosphate in shCtrl adipocytes, whereas intracellular free glucose concentrations were similar to Bnip3 KD (Fig. 5F). Moreover, maximal insulin-stimulated 2-DG uptake in shCtrl cells was more sensitive to ATP synthase inhibition (Fig. 5G), strengthening the link between Bnip3-induced changes in adipose mitochondrial bioenergetics and glucose metabolism.
Finally, we exploited the fact that shB3_1 targets the 3′ untranslated region of Bnip3 mRNA, permitting reconstitution with inducible Bnip3 deletion mutants (Fig. 5H). Interestingly, enforced expression of WT or a mitophagy-deficient Bnip3 (∆LIR) protein reinstated insulin-stimulated 2-DG transport to the level measured in shCtrl adipocytes (Fig. 5I). By contrast, a mutant Bnip3 deficient for mitochondrial targeting (∆TMD) mitigated 2-DG uptake even further, possibly because of a dominant-negative effect (35,36). In addition, expression of WT Nix was unable to compensate for Bnip3 KD (Fig. 5I). These results strongly suggest that mitochondrially targeted Bnip3 improves adipocyte glucose metabolism independent of its promitophagic activity.
Pharmacological Dynamin-Related Protein 1 Inhibition Mimics Bnip3 KD in 3T3-L1 Adipocytes
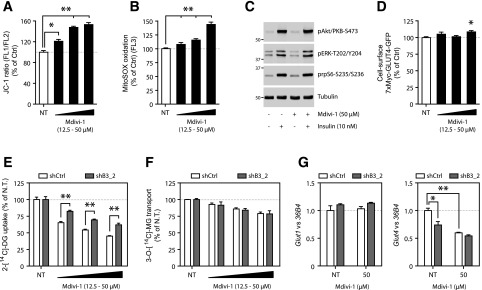
Mitochondrial fission underlies the pathogenesis of obesity-related skeletal muscle IR (13–15). Conversely, adipocytes adopt a fragmented mitochondrial state during differentiation or in response to synthetic PPARγ agonism (9–11). This led us to explore whether the pro-fission activity of Bnip3 is mechanistically required for adipose insulin sensitization. To this end, we incubated 3T3-L1 cells with Mdivi-1, a recently described pharmacological inhibitor of fission/dynamin-related protein 1 (Drp1) (37). Interestingly, Mdivi-1 was previously reported to ameliorate skeletal muscle IR in vitro and in vivo (15).
Short-term Mdivi-1 treatment replicated the effects of Bnip3 KD on adipocyte mitochondrial bioenergetics (Fig. 6A and B). Remarkably, despite normal phosphoinositide 3-kinase/Akt signaling and GLUT4 exocytosis (Fig. 6C and D), Mdivi-1 triggered a dramatic dose-dependent reduction of insulin-mediated 2-DG uptake in 3T3-L1 adipocytes (Fig. 6E). Moreover, Bnip3 KD blunted the efficacy of Mdivi-1 to suppress 2-DG disposal for all concentrations tested (Fig. 6E), suggesting an overlapping mode of action. This insulin-resistant state induced by chemical Drp1 inhibition could not be ascribed to altered Glut4 transcription (data not shown). Similar to Bnip3 KD, Mdivi-1 was less potent in suppressing 3-MG uptake in response to insulin (Fig. 6F). More importantly, the differential effect of Mdivi-1 on 2-DG disposal in shCtrl and shB3_2 adipocytes was not seen for the 3-MG analog (Fig. 6F). This affirms the concept that the insulin-sensitizing action of Bnip3 pro-fission activity is mediated in part by an increased rate of glucose phosphorylation.
Figure 6.
Pharmacological Drp1 inhibition phenocopies the effects of Bnip3 KD in 3T3-L1 adipocytes. Flow cytometric analysis of ∆ψm with JC-1 (A) and mROS formation with MitoSOX (B) in the presence or absence of the mitochondrial fission inhibitor Mdivi-1 (12.5–50 µmol/L) for 5 h. Ctrl, control; NT, not treated. C: Phospho-Akt (pAkt)/PKB, phospho-ERK1/2 (pERK), and phospho-rpS6 (prpS6) levels of 3T3-L1 adipocytes treated with and without Mdivi-1 (50 µmol/L) for 5 h and during the final 15 min with insulin (10 nmol/L). D: Flow cytometric analysis of 7XMyc-GLUT4 plasma membrane occupancy (FL2) in 3T3-L1 adipocytes stably transduced with 7XMyc-GLUT4-GFP, starved (5 h) in the presence or absence of Mdivi-1 (12.5–50 µmol/L), and stimulated with insulin (10 nmol/L) for 15 min. Insulin-stimulated (10 nmol/L) 2-DG uptake (E) and 3-MG transport (F) in the presence of Mdivi-1 (12.5–50 µmol/L) for 5 h. The NT values are expressed as 100% for both shCtrl and shB3_2 to address the relative sensitivity to Mdivi-1. G: Relative Glut1 and Glut4 mRNA levels in 3T3-L1 adipocytes treated with and without Mdivi-1 (50 µmol/L) for 24 h. Values are expressed as mean ± SEM (n = 3 or 4 in A, B, and E–G; n = 6 in D). *P < 0.05; **P < 0.01.
Finally, we aimed to identify the long-term transcriptional effects of enforcing mitochondrial fusion in 3T3-L1 adipocytes by analyzing glucose transporter expression. Overnight Mdivi-1 incubation abrogated Glut4 transcription, whereas Glut1 was unaltered (Fig. 6G). Moreover, Glut4 mRNA levels in shCtrl and shB3_2 adipocytes normalized in the presence of Mdivi-1 (Fig. 6G). Together, these results suggest a mechanistic link between adipocyte mitochondrial network fragmentation and insulin sensitivity, and define Bnip3 as a key regulator of this process.
Adipose-Specific and Systemic Metabolic Adaptations in Bnip3-Deficient Mice
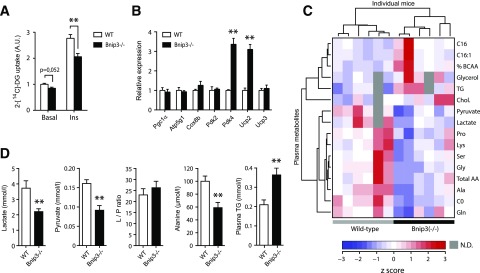
To assess the consequence of altered mitochondrial dynamics on fat cell biology and systemic metabolism, we subjected WT and Bnip3−/− mice to an LF or HF diet. No obvious metabolic and histological abnormalities were observed in Bnip3−/− mice fed an LF diet (Supplementary Fig. 3). In addition, gonadal fat pad weights, adipose cell diameter, histology, and plasma leptin concentrations were all normal in Bnip3−/− mice (Supplementary Fig. 4A–D). Yet, despite similar gWAT mass and function, insulin-induced 2-DG uptake was significantly reduced in gonadal Bnip3−/− versus WT adipocytes (Fig. 7A). Quantitative PCR analysis of gWAT revealed no major alterations in mRNA levels of multiple adipogenic and mitochondrial markers (Supplementary Fig. 4E). Of interest, we observed a robust and selective induction of uncoupling protein 2 (Ucp2) and pyruvate dehydrogenase kinase 4 (Pdk4) in Bnip3−/− gWAT (Fig. 7B). Ucp2 mRNA levels were also markedly elevated in beige and brown fat depots but not in liver and skeletal muscle (Supplementary Fig. 4F), pointing to an adipose-specific compensation for the loss of Bnip3.
Figure 7.
Adipogenic and systemic metabolic reprogramming in Bnip3−/− mice. A: Basal and insulin-stimulated (100 nmol/L) 2-DG uptake in isolated primary gonadal adipocytes. B: Relative mRNA levels of adipogenic and mitochondrial markers in gWAT of WT (white bars) and Bnip3−/− (black bars) mice fed an LF diet for 16 weeks (see also Supplementary Fig. 4G). C: Unsupervised hierarchical clustering of normalized targeted metabolomics on blood and plasma samples of fasted WT and Bnip3−/− mice. Color in the heatmap reflects global metabolite abundance level according to z-score. The top 16 metabolites that contributed to the separation are shown. D: Individual metabolites are highlighted. L/P ratio, lactate-to-pyruvate ratio. Values are expressed as mean ± SEM (n = 6 in A–D; n = 3–4 in B). **P < 0.01 for Bnip3−/− versus WT.
To further outline the systemic effects of Bnip3 deletion, we conducted targeted quantitative metabolomics in the blood of WT and Bnip3−/− cohorts. This led to the identification of a divergent metabolite cluster specifically related to pyruvate and amino acid metabolism (Fig. 7C). The levels of all these glucogenic and glyceroneogenic precursors were significantly lowered in Bnip3−/− mice (Fig. 7C and D). Conversely, we found elevated TG concentrations in Bnip3−/− mice (Fig. 7C and D), indicative of increased hepatic lipogenesis. Taken together, these data suggest that Bnip3 plays a key role in adipose glucose disposal and systemic metabolic regulation.
Bnip3 Deficiency Predisposes Mice to Obesity-Associated IR
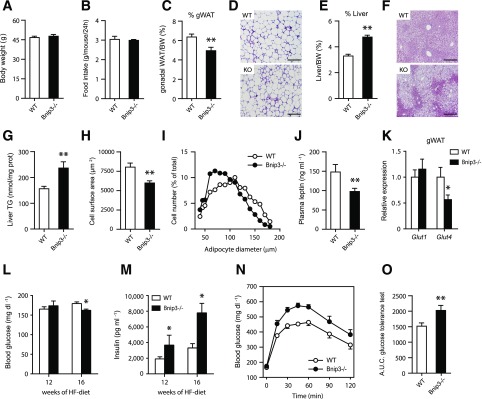
After 16 weeks of HF feeding, Bnip3−/− mice showed no abnormalities in terms of BW gain and food intake relative to WT controls (Fig. 8A and B and Supplementary Fig. 5A). Notably, analysis of BW composition showed that Bnip3−/− mice trade reduced adiposity for increased hepatic steatosis (Fig. 8C–F), which was affirmed histologically and by enhanced liver TG content (Fig. 8F and G). In agreement, we observed attenuated AMPK and acetyl-CoA carboxylase (1/2) phosphorylation in obese Bnip3−/− liver homogenates (Supplementary Fig. 5B), favoring lipid deposition. Conversely, quantitative PCR analysis showed no major changes in gene expression of the hepatic lipogenic program (Supplementary Fig. 5C).
Figure 8.
Bnip3 deficiency predisposes mice to diet-induced insulin resistance. A: BW gain (see also Supplementary Fig. 5A and B). Food intake among WT (white bars) and Bnip3−/− (black bars) mice fed an HF diet for 16 weeks. C and D: gWAT weight (percentage of BW) (C) and hematoxylin-eosin (H-E) staining (D) in WT and knockout mice. E–G: Liver weight (percentage of BW) (E), H-E staining (F), and hepatic TG content (G). Determination of gWAT cell size (H) and diameter distribution (I). J: Analysis of circulating leptin concentrations. K: Relative Glut1 and Glut4 mRNA levels in gWAT (n = 6). L and M: Fasting (4 h) blood glucose (L) and plasma insulin concentrations (M) after 12 and 16 weeks of HF feeding. N and O: Oral glucose tolerance test (2.0 g/kg) in WT and Bnip3−/− mice (N) and analysis of the area under the curve (A.U.C.) (O). Values are expressed as mean ± SEM (n = 7 or 8 in A–O, unless stated otherwise). Scale bars, 300 μm. *P < 0.05, **P < 0.01 for Bnip3−/− versus WT.
On the other hand, Bnip3−/− gWAT showed less hypertrophy after HF feeding, as reflected by decreased fat cell size and diameter relative to WT gWAT (Fig. 8H and I). In line with reduced adiposity, Bnip3−/− mice had lower circulating concentrations of leptin (Fig. 8J). This was not accompanied by altered abundance of plasma TGs and FFAs (Supplementary Fig. 5D and E). In addition, lipogenic and inflammatory mRNA levels in gWAT were similar between the genotypes (Supplementary Fig. 5F). Of note, Glut4 transcription was markedly downregulated in Bnip3−/− gWAT (Fig. 8K). Cumulatively, these findings are all consistent with impaired glucose disposal in gWAT as a mechanism for reduced lipid storage.
We next studied the impact of Bnip3 deficiency on systemic glucose and insulin homeostasis. Surprisingly, fasting (4 h) blood glucose concentrations were normal, or perhaps even slightly decreased, in Bnip3−/− mice (Fig. 8L). Yet this required two- to threefold higher plasma insulin concentrations (Fig. 8M), indicating aggravated systemic IR. Hyperinsulinemia was apparent after 12 weeks of HF feeding and progressed until the point of study termination (Fig. 8M). In addition, Bnip3−/− mice were severely glucose intolerant, as reflected by a ∼25% increase in the area under the curve on glucose tolerance testing (Fig. 8N and O). Finally, we harvested gWAT, liver, and skeletal muscle from weight-matched Bnip3−/− and WT cohorts after intraperitoneal injection with insulin and probed the degree of Akt/protein kinase B (PKB) activation. As shown in Supplementary Fig. 5G–I, Akt/PKB phosphorylation was selectively impaired in obese Bnip3−/− liver, suggesting increased hepatic IR. In aggregate, these results uncover an obesity-driven phenotype of perturbed whole-body glucose and lipid metabolism in Bnip3-null mice.
Discussion
Numerous studies have shown that PPARγ agonism elicits dramatic changes in mitochondrial biogenesis, content, remodeling, respiratory capacity, and fuel selection in gWAT (3–6,8–11). Whether this is required for adipose insulin sensitization and how PPARγ regulates the fusion-fission balance have thus far not been fully explored. By using a combination of mouse population genetics and functional assays, we report here a novel PPARγ-Bnip3 axis that controls mitochondrial network fragmentation in adipocytes. Bnip3 mimics several aspects of PPARγ agonism through its pro-fission activity, such as improved adipose mitochondrial bioenergetics, FA utilization, and insulin-stimulated glucose disposal. Further supporting its metabolic role, Bnip3−/− mice displayed aberrant nonadipose lipid accumulation and exacerbated insulin resistance in response to HF feeding.
A key conceptual advance of this study is that mitochondrial network fragmentation causes adipose tissue to become more insulin sensitive, in contrast to skeletal muscle (15). This differential fusion-fission regulation with respect to insulin sensitivity might be intimately linked to oxidative fuel preference. In the postprandial state, myocytes oxidize glucose in fused/polarized mitochondria, whereas fat cells use glucose mainly for lipogenic purposes and commit to FAO in fragmented/less efficiently coupled mitochondria for ATP synthesis (2). We have shown that enforced fusion in cultured adipocytes, either by Bnip3 KD or Mdivi-1 treatment, inhibits Glut4 transcription. Indeed, mitochondrial catabolism of long-chain FAs has previously been shown to regulate adipose Glut4 expression, most likely by producing ligands of adipocyte-specific transcription factors (38). These data illustrate a tissue-specific interplay between fusion-fission dynamics, nutrient utilization, and insulin sensitivity.
Of note, the ratio of electrons entering the respiratory chain via FADH2 or NADH (F/N) has been proposed as a key determinant in the generation of mROS (39). This may explain why FA breakdown, yielding a much higher F:N ratio than glucose catabolism, triggers elevated radical formation. Intriguingly, although adipose induction of Bnip3 greatly increases the rate of EX-sensitive respiration, this is accompanied by an unexpected reduction in mROS. We propose that a shift toward mitochondrial network fragmentation and increased inherent proton leak may comprise an adaptive metabolic response to protect against high F:N ratios associated with FAO. This is supported by our results showing that enforced mitochondrial elongation in cultured adipocytes led to heightened ∆ψm and FA-induced oxidative damage. In addition, Ucp2 was selectively upregulated in murine Bnip3−/− fat depots. By eliciting mild uncoupling of OXPHOS during FAO (40), Ucp2 may suppress radical formation in “inappropriately” fused mitochondrial networks in Bnip3−/− adipose tissues. Lastly, inadequate fission may also underlie exacerbated ROS levels in overnight-fasted Bnip3−/− livers (41).
Glick et al. (41) were the first to report a major role for Bnip3 in the regulation of hepatic lipid metabolism. Bnip3 protein levels are robustly upregulated during the hepatic fasting response, and loss of Bnip3 leads to impaired FAO and the development of severe liver steatosis. Of note, although initially ascribed to defective mitophagy, more recent evidence from the same laboratory showed that Bnip3 also modulates FAO in a manner distinct from its promitophagic activity (42). This is in line with our findings showing that the Bnip3-dependent increase in adipose FAO occurred independent of changes in mitochondrial mass and turnover. Whether Bnip3-regulated fission is sufficient for optimal FAO requires further investigation. It is tempting to speculate that fragmentation results in an extension of the mitochondrial–cytoplasmic interface, enabling an accelerated rate of FA import and FAO. Supporting evidence that fission/mitophagy are genetically linked to FAO comes from studies aimed to identify novel AMPK substrates. In addition to its known role in stimulating FAO, AMPK promotes fission and mitophagy by directly phosphorylating the mitochondrial DRP1-receptor MFF (43) and the autophagy protein ULK1 (44). Bnip3 may play a complementary role by inhibiting the core fusion machinery (17,18) and serving as a mitophagy receptor (20–22) to ensure optimal mitochondrial function during FAO. Notably, it has been shown that Fis1 overexpression in a liver-specific murine model of dysregulated fusion-fission dynamics ameliorates liver steatosis, supporting the relevance of proper fission in hepatic lipid metabolism (45).
Relatively few studies to date have addressed the functional role of fusion-fission processes in adipocytes. Kita et al. (10) showed that mitochondrial network fragmentation improves triacylglycerol accumulation in 3T3-L1 adipocytes. Consistent with this, observations made in two independent mouse models and in cultured cells indicate that Bnip3 expression positively associates with adipose storage capacity. Indeed, fission may enhance glucose incorporation into lipids by facilitating accelerated export of the Krebs cycle intermediate citrate. Of interest, genes related to fission (Bnip3, Drp1) were robustly downregulated in gWAT of murine models of dietary and genetic obesity, whereas those involved in fusion (Mfn1/2, Opa1) were only marginally affected. Conversely, an opposite trend was detected in brown adipose tissue (BAT), with elevated expression of pro-fission genes in the obese state. The reciprocal regulation of Bnip3 expression in white and brown fat depots may reflect an adaptive metabolic response to cope with excess dietary lipids, that is, by limiting further lipid deposition in gWAT and enhancing FA utilization in BAT depots.
We have shown here that Bnip3−/− mice have normal adipose mass despite blunted insulin-mediated glucose uptake in gWAT. Under conditions of glucose scarcity, white adipocytes rely strictly on glyceroneogenesis for glycerol-3-phosphate generation (46). Interestingly, Bnip3−/− mice exhibit lower concentrations of plasma pyruvate, lactate, and alanine, the main precursors of the glyceroneogenic pathway. Moreover, Pdk4 and Ucp2 are highly upregulated in Bnip3−/− gWAT. Pdk4 has been reported to stimulate glyceroneogenesis in gWAT by acting as a pyruvate-sparing switch via its ability to suppress the pyruvate dehydrogenase complex (47). Accordingly, by transporting C4 metabolites out of mitochondria (48), Ucp2 limits pyruvate conversion to acetyl-CoA and concurrently raises cytosolic concentrations of malate and oxaloacetate, which could be used for glycerol-3-phosphate synthesis. Although hitherto not formally linked to glyceroneogenic control in WAT, Ucp2 expression is induced upon HF feeding or TZD administration (40), known activators of the glyceroneogenic pathway. Whether Bnip3−/− fat depots indeed show enhanced glyceroneogenic activity and contribute to the lowering of plasma glucogenic precursors requires further investigation.
A study by Choi et al. (49) validated Bnip3 as a direct transcriptional target of PPARγ in adipose tissue. We substantiate their data and also provide evidence supporting the necessity of C/EBPα in the regulation of Bnip3 expression. Similar to our results, Choi et al. also reported that Bnip3 is critical for adipocyte mitochondrial bioenergetics, but they ascribed this to a Bnip3-related increase in mitochondrial biogenesis (49). We have exhaustively analyzed various indices for mitochondrial content but were unable to find disparities between shCtrl and Bnip3-KD adipocytes. Instead, we have shown here that the Bnip3-dependent increase in OCR was related to altered mitochondrial dynamics and fuel utilization. This apparent discrepancy might be the result of varying experimental conditions, such as the presence of rosiglitazone during analyses (50).
In conclusion, our studies describe a regulatory circuit in which PPARγ controls mitochondrial remodeling through transcriptional induction of Bnip3. Importantly, we further link its pro-fission activity to improved adipose mitochondrial bioenergetics and glucose disposal. Our data highlight that the fusion-fission balance is regulated by tissue-specific processes that could have distinct or even opposing effects with respect to insulin sensitivity. Potential antidiabetic strategies targeting mitochondrial dynamics should therefore act in a tissue-selective manner.
Article Information
Acknowledgments. The authors are grateful to all members of the Aerts laboratory for helpful and stimulating discussions, in particular Dr. A.J. Meijer and Dr. D. Speijer. Dr. C.L. Vrins, Dr. B. Bleijlevens, J. Houben-Weerts, D.S. Koenis, and S. van den Berg are acknowledged for technical assistance. The authors are grateful to Dr. R.H. Houtkooper (Department of Genetic Metabolic Diseases, University of Amsterdam) for assistance with Seahorse experiments and Prof. V. Everts and W. Tigchelaar (Department of Cell Biology, University of Amsterdam) for help with EM studies.
Funding. This work was supported by the Dutch Diabetes Foundation (no. 2009.80.016 to M.v.E.) and the National Institutes of Health (no. HL59888 to G.W.D. and no. DK092661 to J.S.B.). N.Z. is an Established Investigator of the Dutch Heart Foundation (2013T111) and is supported by an ERC Consolidator grant (617376) from the European Research Council.
Duality of Interest. No conflicts of interest relevant to this article have been reported.
Author Contributions. M.J.T. conceived the project, designed and performed most experiments, and wrote the manuscript. R.O. supervised and performed in vivo analyses. M.v.E., J.A., S.M.H., C.v.R., M.C.B., and S.S. researched data. N.Z. contributed to the discussion and critically reviewed and edited the manuscript. N.Z., D.G., J.M.A., J.S.B., and G.W.D. provided key reagents and/or expert advice that enabled key parts of this study. M.A.H. probed published chromatin immunoprecipitation sequencing data. C.A.A. conducted in silico and in vivo studies and critically reviewed and edited the manuscript. A.J.V. supervised the study and critically reviewed and edited the manuscript. A.J.V. is the guarantor of this work and, as such, had full access to all data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0243/-/DC1.
References
- 1.Cohen P, Spiegelman BM. Brown and beige fat: molecular parts of a thermogenic machine. Diabetes 2015;64:2346–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab 2012;23:435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson-Fritch L, Nicoloro S, Chouinard M, et al. . Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 2004;114:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rong JX, Qiu Y, Hansen MK, et al. . Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 2007;56:1751–1760 [DOI] [PubMed] [Google Scholar]
- 5.Bogacka I, Xie H, Bray GA, Smith SR. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes 2005;54:1392–1399 [DOI] [PubMed] [Google Scholar]
- 6.Choo HJ, Kim JH, Kwon OB, et al. . Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 2006;49:784–791 [DOI] [PubMed] [Google Scholar]
- 7.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994;79:1147–1156 [DOI] [PubMed] [Google Scholar]
- 8.Shi X, Burkart A, Nicoloro SM, Czech MP, Straubhaar J, Corvera S. Paradoxical effect of mitochondrial respiratory chain impairment on insulin signaling and glucose transport in adipose cells. J Biol Chem 2008;283:30658–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson-Fritch L, Burkart A, Bell G, et al. . Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 2003;23:1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kita T, Nishida H, Shibata H, Niimi S, Higuti T, Arakaki N. Possible role of mitochondrial remodelling on cellular triacylglycerol accumulation. J Biochem 2009;146:787–796 [DOI] [PubMed] [Google Scholar]
- 11.Ducluzeau PH, Priou M, Weitheimer M, et al. . Dynamic regulation of mitochondrial network and oxidative functions during 3T3-L1 fat cell differentiation. J Physiol Biochem 2011;67:285–296 [DOI] [PubMed] [Google Scholar]
- 12.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab 2013;17:491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 2002;51:2944–2950 [DOI] [PubMed] [Google Scholar]
- 14.Bach D, Pich S, Soriano FX, et al. . Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem 2003;278:17190–17197 [DOI] [PubMed] [Google Scholar]
- 15.Jheng HF, Tsai PJ, Guo SM, et al. . Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol 2012;32:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 2009;16:939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landes T, Emorine LJ, Courilleau D, Rojo M, Belenguer P, Arnauné-Pelloquin L. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep 2010;11:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol 2011;301:H1924–H1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellot G, Garcia-Medina R, Gounon P, et al. . Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009;29:2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Bosch-Marce M, Shimoda LA, et al. . Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008;283:10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 2012;287:19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Massen S, Terenzio M, et al. . Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem 2013;288:1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novak I, Kirkin V, McEwan DG, et al. . Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010;11:45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics 2003;1:299–308 [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Schadt EE, Wang S, et al. . Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res 2006;16:995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, et al. . Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R, Pedersen TA, Hagenbeek D, et al. . Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev 2008;22:2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel TL, Tol MJ, Ottenhof R, et al. . Lysosomal stress in obese adipose tissue macrophages contributes to MITF-dependent Gpnmb induction. Diabetes 2014;63:3310–3323 [DOI] [PubMed] [Google Scholar]
- 29.Meerbrey KL, Hu G, Kessler JD, et al. . The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 2011;108:3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison SA, Buxton JM, Clancy BM, Czech MP. Evidence that erythroid-type glucose transporter intrinsic activity is modulated by cadmium treatment of mouse 3T3-L1 cells. J Biol Chem 1991;266:19438–19449 [PubMed] [Google Scholar]
- 31.Bogan JS, McKee AE, Lodish HF. Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells: regulation by amino acid concentrations. Mol Cell Biol 2001;21:4785–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diwan A, Krenz M, Syed FM, et al. . Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 2007;117:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houten SM, Herrema H, Te Brinke H, et al. . Impaired amino acid metabolism contributes to fasting-induced hypoglycemia in fatty acid oxidation defects. Hum Mol Genet 2013;22:5249–5261 [DOI] [PubMed] [Google Scholar]
- 34.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci 2014;37:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamacher-Brady A, Brady NR, Logue SE, et al. . Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ 2007;14:146–157 [DOI] [PubMed] [Google Scholar]
- 36.Azad MB, Chen Y, Henson ES, et al. . Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 2008;4:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cassidy-Stone A, Chipuk JE, Ingerman E, et al. . Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 2008;14:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griesel BA, Weems J, Russell RA, Abel ED, Humphries K, Olson AL. Acute inhibition of fatty acid import inhibits GLUT4 transcription in adipose tissue, but not skeletal or cardiac muscle tissue, partly through liver X receptor (LXR) signaling. Diabetes 2010;59:800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speijer D. Oxygen radicals shaping evolution: why fatty acid catabolism leads to peroxisomes while neurons do without it: FADH₂/NADH flux ratios determining mitochondrial radical formation were crucial for the eukaryotic invention of peroxisomes and catabolic tissue differentiation. Bioessays 2011;33:88–94 [DOI] [PubMed] [Google Scholar]
- 40.Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab 2005;2:85–93 [DOI] [PubMed] [Google Scholar]
- 41.Glick D, Zhang W, Beaton M, et al. . BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol 2012;32:2570–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boland ML, Huang H, Shah R, et al. BNip3 connects energy sensing to hepatic metabolism and mitophagy [abstract]. Proceedings of the 104th Annual Meeting of the American Assocation for Cancer Research 2014 Apr 5–9; San Diego, CA: AACR; Cancer Res 2014;74(19 Suppl): Abstract nr 4324
- 43.Toyama EQ, Herzig S, Courchet J, et al. . Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016;351:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egan DF, Shackelford DB, Mihaylova MM, et al. . Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011;331:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobi D, Liu S, Burkewitz K, et al. . Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab 2015;22:709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reshef L, Olswang Y, Cassuto H, et al. . Glyceroneogenesis and the triglyceride/fatty acid cycle. J Biol Chem 2003;278:30413–30416 [DOI] [PubMed] [Google Scholar]
- 47.Cadoudal T, Distel E, Durant S, et al. . Pyruvate dehydrogenase kinase 4: regulation by thiazolidinediones and implication in glyceroneogenesis in adipose tissue. Diabetes 2008;57:2272–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vozza A, Parisi G, De Leonardis F, et al. . UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proc Natl Acad Sci U S A 2014;111:960–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi JW, Jo A, Kim M, et al. . BNIP3 is essential for mitochondrial bioenergetics during adipocyte remodelling in mice. Diabetologia 2016;59:571–581 [DOI] [PubMed] [Google Scholar]
- 50.Brunmair B, Staniek K, Gras F, et al. . Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes 2004;53:1052–1059 [DOI] [PubMed] [Google Scholar]