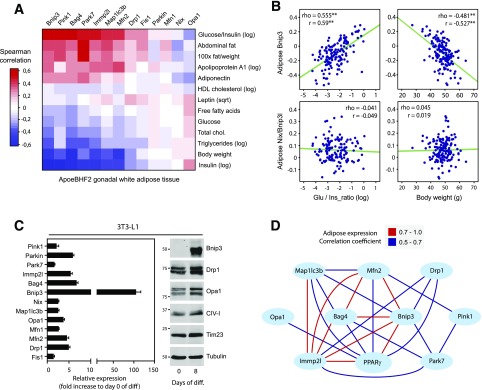
Figure 1.
Identification of Bnip3 as a potential PPARγ effector regulating adipose mitochondrial dynamics. A: Heatmap transformation of gene/trait correlations for mitochondrial dynamics–related mRNA transcripts in gWAT versus the indicated metabolic phenotypes scored across individual males from the BxH apolipoprotein E null F2 mouse cross (BHF2; n = ∼140). The axes were hierarchically clustered (Euclidean distance). B: Correlation matrix plots between adipose Bnip3 and Nix transcript levels and phenotypic traits, glucose-to-insulin ratio, and BW (r = Pearson and rho = Spearman). C: Quantitative PCR and immunoblot analyses of mitochondrial dynamics–related expression in 3T3-L1 adipocytes (n = 4). D: Pparγ mRNA expression shows covariance with mitochondrial dynamics–related transcript levels in gWAT. Red lines indicate a positive Pearson correlation coefficient of 0.7–1.0, and blue lines signify a coefficient of 0.5–0.7. Phenotypic trait nomenclatures are as follows: abdominal fat (grams), cholesterol (milligrams per deciliter), FFAs (milligrams per deciliter), HDL fraction of cholesterol log (milligrams per deciliter), glucose (milligrams per deciliter), plasma insulin log (picograms per liter), TGs log (milligrams per deciliter). diff, differentiation; glu, glucose; ins, insulin; log, log transformed; sqrt, square root transformed. **P < 0.01.