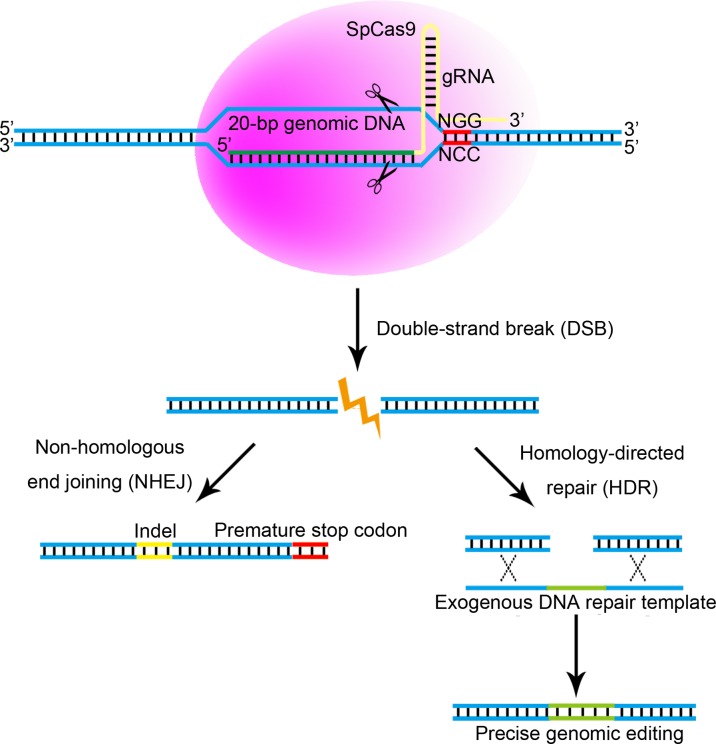
Figure 2.
The CRISPR-Cas9 system. The Type II Streptococcus pyogenes clustered regulatory interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) (SpCas9) system, which is the simplest and most extensively used CRISPR-Cas9 technology, is based on a guide-RNA (gRNA) containing a specific 20 bp sequence to guide the DNA endonuclease Cas9 to a complementary target DNA sequence in the genome where it induces a DNA double-strand break (DSB). The 20-bp target genomic DNA must be upstream of a specific sequence (5′-NGG, where N represents a random nucleotide). The Cas9-induced DSB occurs ~3-bp upstream of the 5′-NGG, and can in theory be induced in any 20-bp genomic DNA sequence flanking 5′-NGG. The Cas9-induced DSB will then be repaired by either homology-directed repair (HDR), which can occur with the presence of DNA repair templates, or by non-homologous end joining (NHEJ). The error-prone NHEJ creates insertions/deletions (indels) around the DSB point. Indels, especially when occurring in early coding exons, can cause loss of gene function (gene knockout) by causing a frame shift that can lead to formation of a pre-mature stop codon. In contrast, HDR uses a template sequence for very precise repair of the DSB. Exogenous DNA repair templates (with the required sequences placed between homology arms) can be provided to the cells together with other components of the CRISPR-Cas9 system to create specific indels or modifications at target genomic loci. Thus, the CRISPR-Cas9 system can be used to insert sequences or correct disease-causing mutations in a very accurate way.