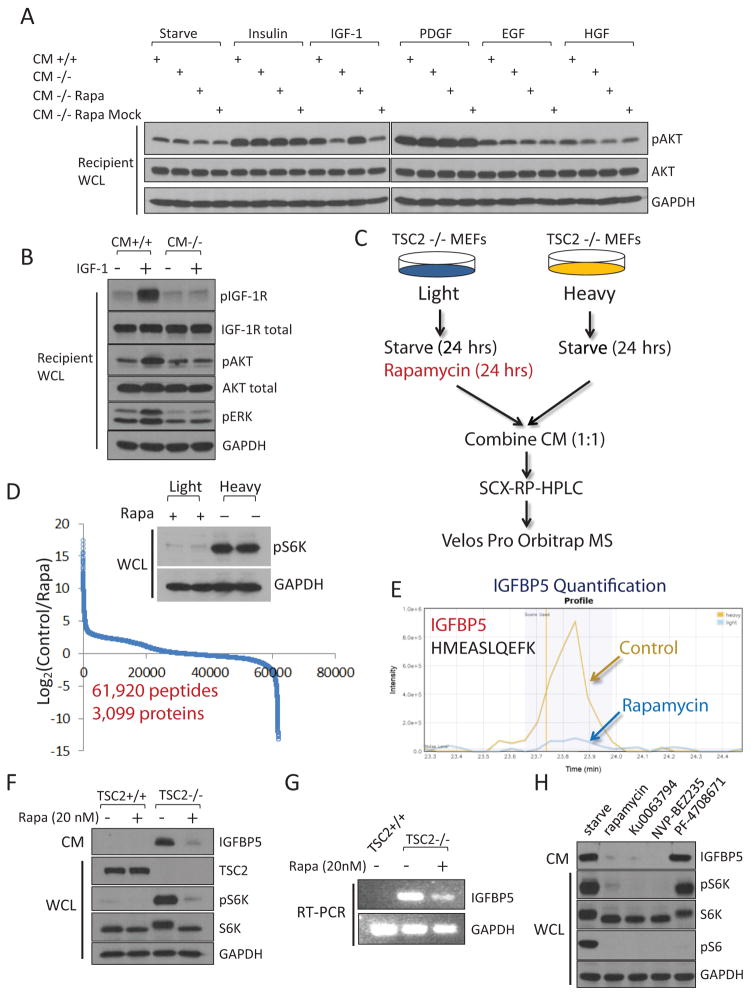
Figure 1.
Cells with hyperactivated mTORC1 secrete a protein factor(s) that blocks IGF-1 signaling. (A) Conditioned media was collected from TSC2+/+ MEFs (CM+/+) or TSC2−/− MEFs (CM−/−), mixed with the indicated growth factor, and were then incubated with regular wt MEFs (designated as “recipient cells”) for 10 min. CM that was not mixed with any growth factors was indicated as “starve”. CM was also collected from TSC2−/− MEFs that had been treated with 20 nM rapamycin for 24 hrs (CM−/− Rapa). As a control experiment, CM from TSC2−/− cells were collected first, and then mixed with rapamycin (CM−/− Rapa Mock). For site-specific phosphorylation, pAkt(S473) levels were analyzed. Growth factor concentrations are Insulin, 100 nM; IGF-1, 40 ng/ml; PDGF, 50 ng/ml; EGF, 50 ng/ml and HGF, 50 ng/ml. (B) CM from TSC2−/− MEFs is able to block the activation of the IGF-1 signaling pathway. Experiments were performed as in (A). pIGF-1R(Y1135/1136), pAkt(S473), and pERK(T202/Y204) levels were analyzed. (C) A gerneral schematic of the quantitative secretomic platform. (D) Ratio distribution of the identified peptides (a total of 63,157 from 3,430 proteins). Ratio (control/rapamycin-treated) distribution of these peptides is shown on a Log2 scale. Light and heavy lysates were also subject to immunoblotting analysis for pS6K(T389) levels. (E) Extracted ion chromatogram of the light (rapamycin-treated, blue) and heavy (control, yellow) ions of an IGFBP5 peptide (HMEASLQEFK). (F) CM from TSC2−/− MEFs, but not TSC2+/+ MEFs, contains high levels of IGFBP5. Cells were starved for 24 hrs, after which CM was collected. When indicated, cells were also treated with rapamycin (20 nM for 24 hrs). CM and WCL of these cells were analyzed by using immunoblotting experiments for pS6K(T389) levels. (G) IGFBP5 expression is regulated by mTORC1 at the transcription level. Total RNA was extracted from TSC2+/+, TSC2−/− MEFs, or TSC2−/− MEFs that had been treated with 20 nM rapamycin for 24 hrs, and was analyzed. (H) Treatment of TSC2−/− MEFs by rapamycin (20 nM), Ku0063794 (1 μM) and NVP-BEZ235 (500 nM), but not an S6K inhibitor (PF-4708671, 10 μM), led to downregulation of IGFBP5 in CM. For site-specific phosphorylation, pS6K(T389) and pS6(S235/236) levels were analyzed.