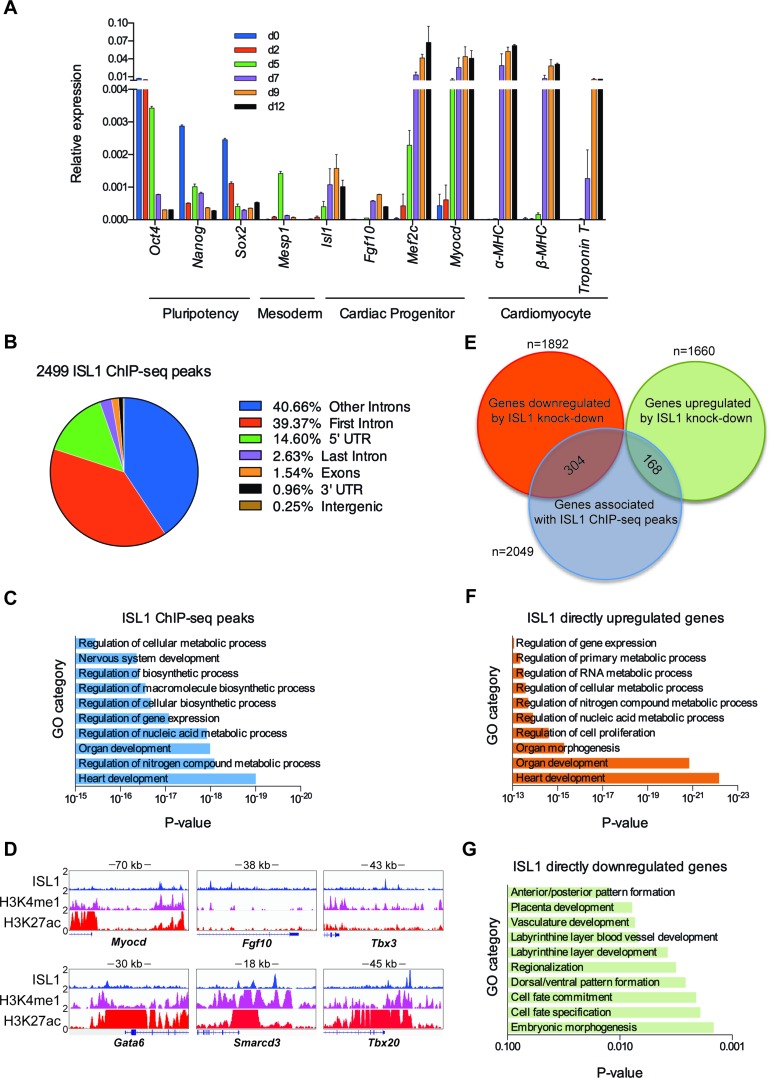
Figure 1.
Genome-wide downstream targets of ISL1 during cardiac progenitor differentiation. (A) Relative mRNA expression of pluripotency (Oct4, Sox2 and Nanog), mesoderm (Mesp1), cardiac progenitor (Isl1, Fgf10, Mef2c and Myocd) and cardiomyocyte (α-MHC, β-MHC and Troponin T) markers in cardiac differentiation of ESCs was measured by qRT-PCR at indicated time points. 18S rRNA was used as an internal control. Data are mean ± SD, n = 4. (B) ChIP-seq ISL1-binding regions were mapped relative to their nearest downstream genes. Annotation includes whether a peak is in the first intron, other introns, 5′ UTR, last intron, exons (coding), 3′ UTR, intergenic. (C) GO functional clustering of genes associated with ISL1 ChIP-seq peaks (top 10 categories are shown). (D) The binding of ISL1 in EBs at day 7 of cardiac differentiation, H3K4me1 and H3K27ac in embryonic day 14.5 of heart tissue on representative ISL1 target genes, Myocd, Fgf10, Tbx3, Gata6, Smarcd3 and Tbx20. (E) Overlay of RNA-seq and ChIP-seq results revealed 472 genes as potential direct targets of ISL1 in day 7 EBs of differentiation. (F and G) GO functional clustering of genes allowed for identification of cellular functions directly regulated by ISL1 (top 10 categories are shown).