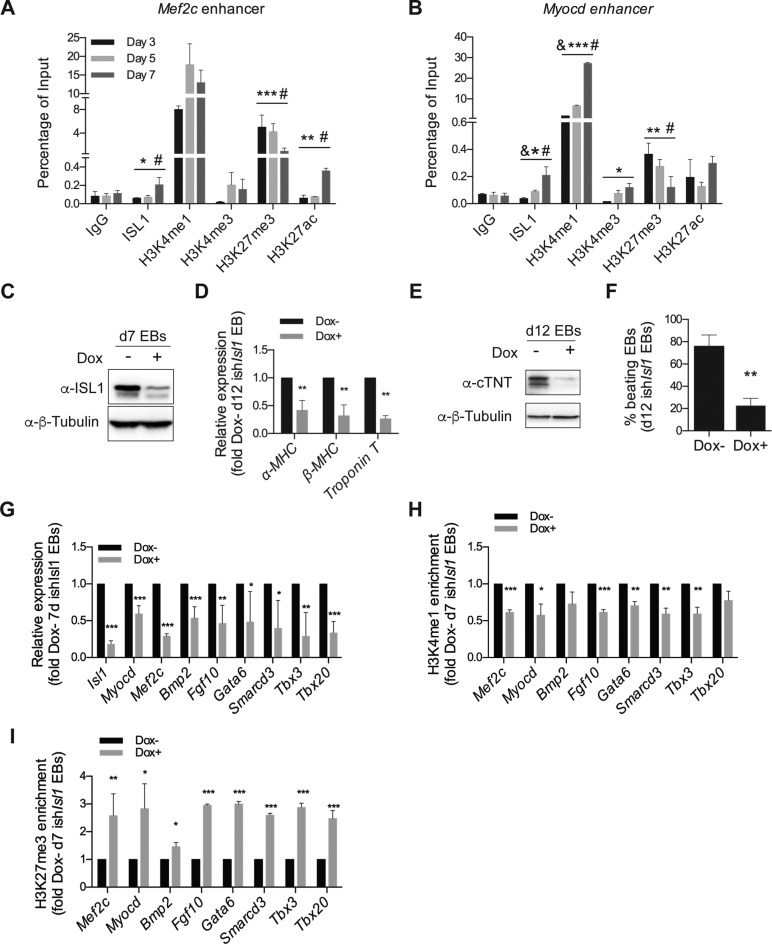
Figure 2.
ISL1 perturbation impairs demethylation of H3K27me3 at the enhancers of its downstream cardiac-specific target genes. (A and B) ChIP of nuclear extracts from day 3, 5 and 7 EBs differentiated from ESCs using anti-ISL1, anti-H3K4me1, anti-H3K4me3, anti-H3K27me3 and anti-H3K27ac or IgG as the control. qRT-PCRs were performed using primers flanking conserved ISL1 binding sites in the Mef2c enhancer (A) and the Myocd enhancer (B). Data are mean ± SD, n = 3. &P < 0.01 d5 VS d3, *P < 0.05 d7 versus d3, **P < 0.01 d7 versus d3, ***P < 0.001 d7 versus d3, #P < 0.01 d7 versus d5. (C) Western blot analysis of total protein extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. β-Tubulin served as a loading control. (D) Relative mRNA expression of cardiomyocyte genes (α-MHC, β-MHC and Troponin T) in day 12 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (E) Western blot analysis of total protein extracts of day 12 EBs differentiated from ishIsl1 in the presence or absence of Dox. β-Tubulin served as a loading control. (F) Percentage of beating EBs in day 12 EBs differentiated from ishIsl1 in the presence or absence of Dox. (G) Relative mRNA expression of Isl1 and its downstream target genes (Myocd, Mef2c, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20 of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox. (H and I) ChIP of nuclear extracts of day 7 EBs differentiated from ishIsl1 ESCs in the presence or absence of Dox using anti-H3K4me1 (H) and anti-H3K27me3 (I). qRT-PCRs were performed using primers targeting ISL1 binding sites in the enhancers of Mef2c, Myocd, Bmp2, Fgf10, Gata6, Smarcd3, Tbx3 and Tbx20. ChIP enrichments are normalized to Input and are represented as fold change relative to Dox- EBs. Data in D and F-I are mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.