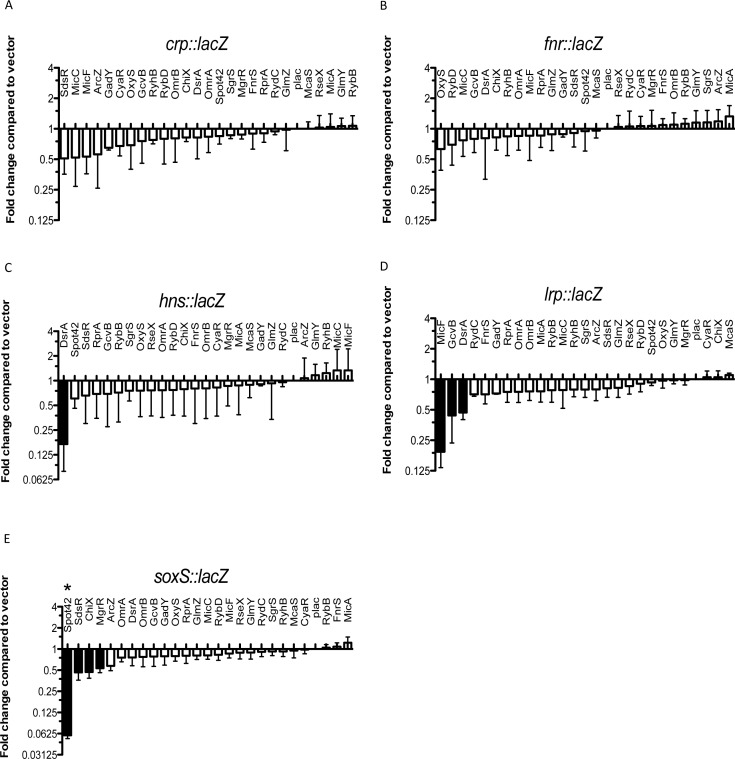
Figure 1.
Use of a library of small RNAs to study translational regulation of transcription factors. Screening of the sRNA library on the PBAD-crp-lacZ fusion (HL1070) (A), the PBAD-fnr-lacZ fusion (HL1069) (B), the PBAD-hns-lacZ fusion (HL1061) (C), the PBAD-lrp-lacZ fusion (HL1044) (D) and the PBAD-soxS-lacZ fusion (HL1064) (E). Cells were grown on LB medium containing 100-μg/ml ampicillin, 100-μM IPTG and 0.001% arabinose (for soxS-lacZ fusion; 0.0001% arabinose). The effect of the overexpression of each sRNA on each fusion was plotted as a function of the fold change compared with the basal activity of the same fusion containing a pBRplac control vector. Basal specific activities for each fusion (in machine units, about 25x less than Miller units) are 145 units for crp; 90 units for fnr, 1899 units for hns, 314 units for lrp and 352 units for soxS. Fold changes greater than two were considered significant (black bars). Assays were done in quadruplicate; error bars indicate standard deviation. * Spot 42 has been found to block arabinose induction of the pBAD promoter at the very low arabinose concentrations used for the soxS fusion (J. Chen, in preparation).