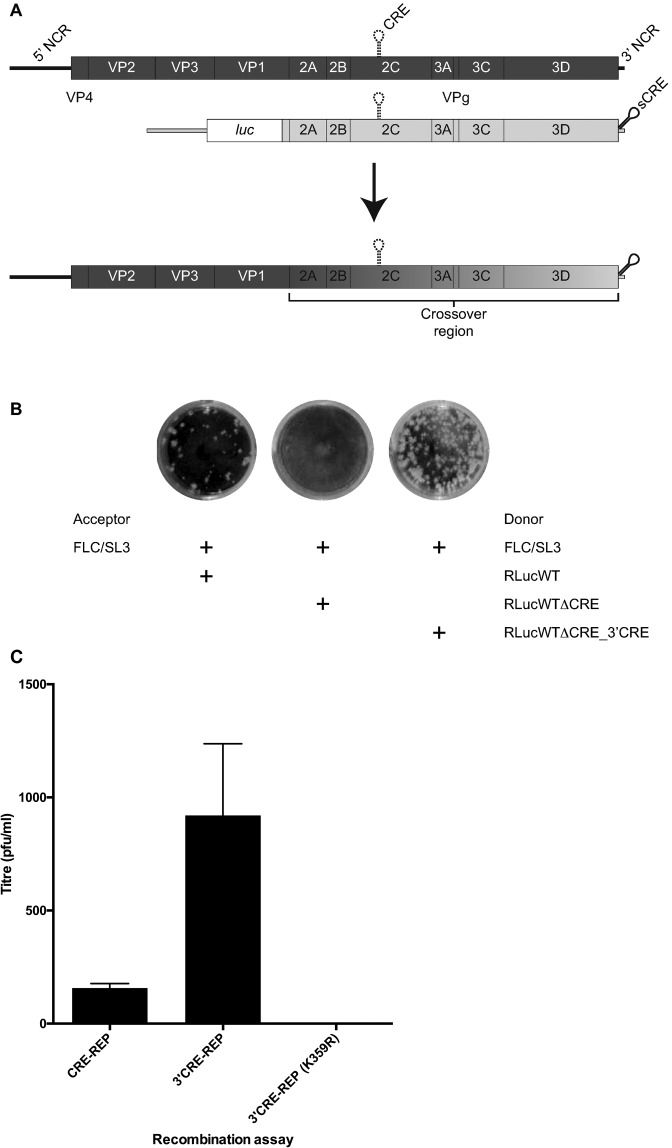
Figure 3.
An extended 3′CRE-REP recombination assay. (A) Schematic depiction of the 3′CRE-REP recombination assay. The acceptor genome (dark shading) bearing a defective CRE indicated as a broken line in the 2C-coding region is shown above a representation of the donor genome, a luciferase-encoding sub-genomic replicon (light shading) in which the native CRE has been inactivated and a synthetic CRE (indicated sCRE) inserted into the 3′ NCR. Following co-transfection of permissive cells (indicated by an arrow), a replication competent recombinant genome may be recovered of the generic structure shown, consisting of the 5′ part of the genome derived from full-length, capsid-encoding, acceptor genome and the 3′ part from the luciferase-encoding donor replicon. The crossover may occur within the region indicated. (B) Increased recombinant yields from the 3′CRE-REP assay. 250 ng of the acceptor or each of the indicated donor RNAs were co-transfected into L929 murine cells and supernatant harvested at 48 h post-transfection. Recombinant virus present in similar dilutions of supernatant was compared by plaque assay in HeLa cells and stained 72 h post infection. (C) Comparison of intratypic recombination yields from CRE-REP and 3′CRE-REP assays primed with RLucWT and FLC/SL3 donor and acceptor genomes. The presence of K359R polymerase mutation in both donor and acceptor genomes prevented the generation of recombinants in the 3′CRE-REP assay. The data represents the mean from three independent samples (± standard deviation). All plaque assay results were confirmed by TCID50 (data not shown).