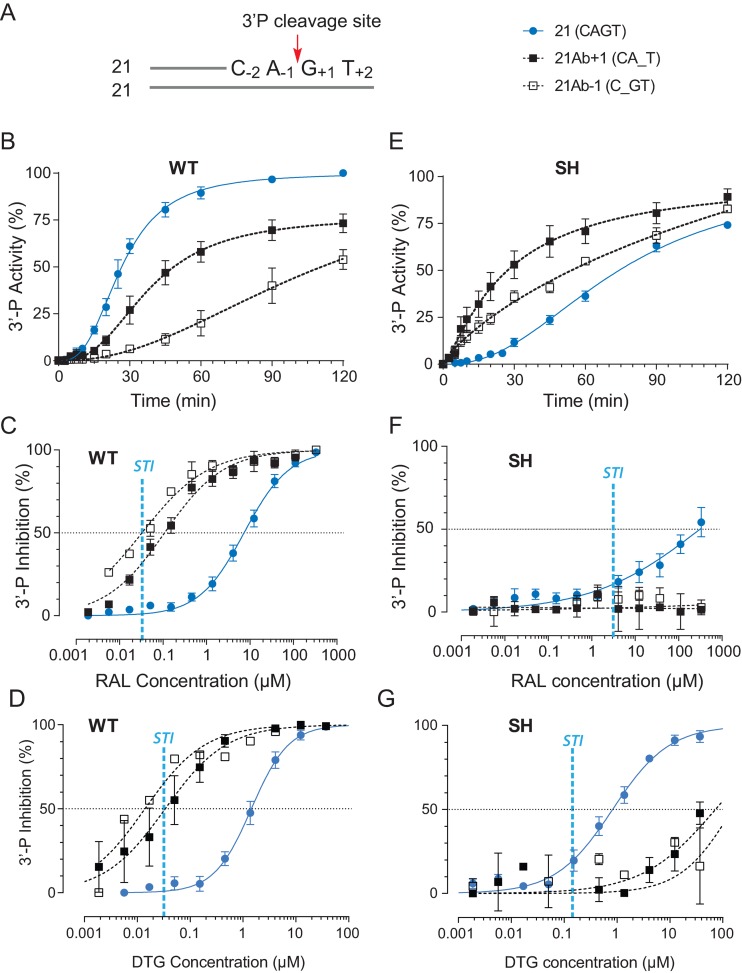
Figure 1.
Differential 3′-processing of modified DNA substrates by the wild-type IN (WT) and the drug-resistant IN mutant G140S-Q148H (SH), and inhibition of 3-processing by raltegravir (RAL) and dolutegravir (DTG). (A) Schematic representation of the 21 base pair duplex oligonucleotide used as IN substrate. The canonical CAGT terminal sequence is numbered according to the 3′-P cleavage site (red arrow). Modified substrates have a tetrahydrofuran in place of a nucleobase at the locations, which is indicated by an underscore. 3′-P activity was monitored with the indicated substrates using WT (B) or the RAL-resistant SH mutant (E). Using the 120-min time point, the inhibition of 3′-P by increasing concentration of RAL or DTG for the various substrates was measured in reactions with WT IN (C and D) or the SH mutant (F and G). Fit curves and standard deviation (SD) were derived from at least 4 and up to 14 independent experiments. For comparison, the vertical blue dashed lines in panels C, D, F and G represent the IC50 value for RAL and DTG for ST inhibition (STI) for WT IN and the SH mutant.