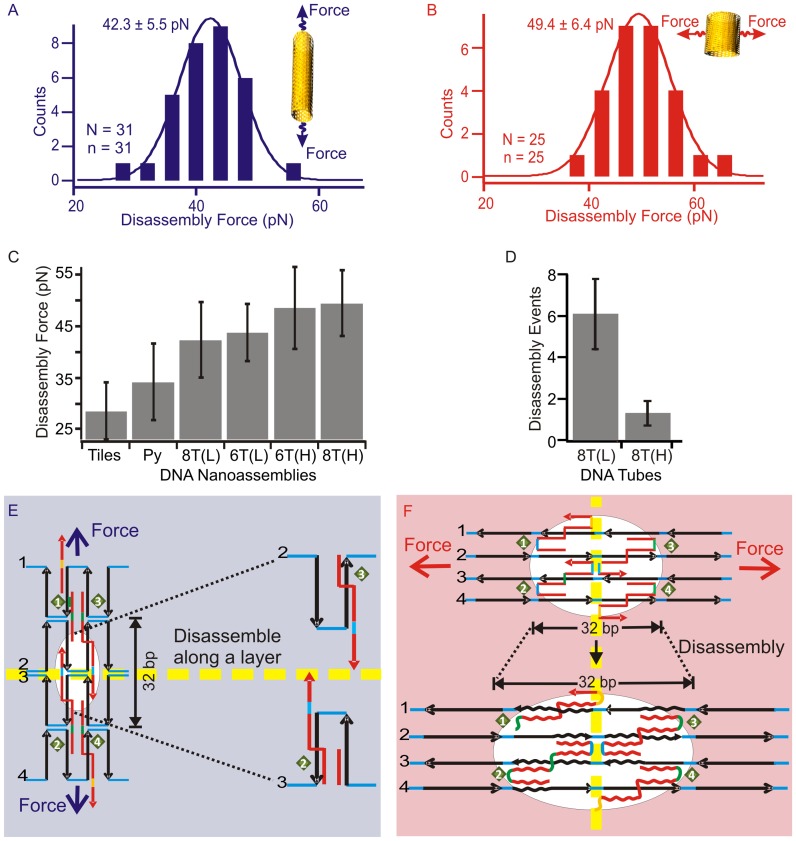
Figure 3.
Disassembly processes of DNA origami structures. (A and B) Histograms of the mechanical disassembly forces measured along longitudinal and horizontal stretching directions of DNA eight-tubes, respectively. The disassembly force was determined at the first disassembly event (the red arrow in Figure 2B). The insets depict the stretching directions. N and n respectively indicate the number of F–X traces and the number of molecules. (C) Comparison of the disassembly force of DNA nanotiles (Tiles), DNA nanopyramid (Py), eight-tube DNA (8T) and six-tube DNA (6T). (D) Average number of the disassembly events observed during the mechanical disintegration of the 8-tube DNA (8T) in longitudinal (L) or horizontal (H) stretching. (E and F) Schematics of the disassembly with respect to Holliday junctions during longitudinal and horizontal stretching directions, respectively. Dotted yellow lines depict Holliday junction layers in different stretching orientations. For clarity, only one complete Holliday junction is shown in the white circle in each scheme.