Abstract
Academic literature provides rich and up-to-date information concerning adverse drug reactions (ADR), but it is time consuming and labor intensive for physicians to obtain information of ADRs from academic literature because they would have to generate queries, review retrieved articles and summarize the results. In this study, a method is developed to automatically detect and summarize ADRs from journal articles, rank them and present them to physicians in a user-friendly interface. The method studied ADRs for 6 drugs and returned on average 4.8 ADRs that were correct. The results demonstrated this method was feasible and effective. This method can be applied in clinical practice for assisting physicians to efficiently obtain information about ADRs associated with specific drugs. Automated summarization of ADR information from recent publications may facilitate translation of academic research into actionable information at point of care.
Introduction
Adverse drug reactions (ADRs) are negative effects on patients resulting from an intervention related to the use of medicines1. It has been considered one of the top issues regarding patient safety and quality of care2,3. ADRs occur commonly in hospitals, and it has been shown that about 1900 of ADRs happen in a single hospital per year on average4 –6. Many of the ADRs are preventable, but still occur because of the lack of up to date ADR knowledge of the physicians in the ordering stage4,7. Due to this situation, dissemination of ADR information at the point of care can be helpful for physicians, and can prevent severe side effects of patients when giving prescriptions, thereby increasing drug safety prospectively8,9 Academic literature provides solid pharmacological information of drugs, which could be helpful in reducing errors related to ADRs. However, it is difficult and time-consuming for physicians to perform a comprehensive review because it requires computational skills to create appropriate queries to retrieve articles discussing ADRs, and it requires physicians to review the articles returned to see if they contain useful ADR information. Both of these tasks are not only time consuming, the overload of information may itself be responsible for errors10. Therefore, a compact display of information with links that provide easy access to the most relevant articles is necessary to help reduce clinical errors and improve patient safety11.
Previous work was published concerning development of methods that aim to extract articles from PubMed that provide information about ADRs. PubMed is an on-line server for MEDLINE database, providing over 24 million citations of biomedicine literature articles18. It also includes databases in different areas of biomedicine. Some articles concerning previous work were only focused on finding drugs associated with a specific ADR12,13. Some previous work used textual features and indexing associated with MEDLINE, and others used machine learning methods12,20. Adams et al. used a method based on customization of PubMED queries to retrieve articles concerning ADRs associated with a specific drug21. Wang et al. used a machine learning method to classify journal articles and determine ADR-drug pairs for a specific ADR13. Shetty et al. used statistical methods to recognize ADR-drug pairs from PubMed literature14. Gurulingappa et al. used an annotated ADR corpus to train a machine learning model to extract potential ADRs from MEDLINE case reports15,16. Some other work involved applications in decision making based on manually generated protocols as input10,17. Manually generated protocols need human labor and cannot be updated in a timely fashion. Our aim is different from previous work in that our method is meant to serve as a tool for physicians to quickly access ADR information discussed in the most relevant articles. Our method processes retrieved articles concerning ADRs further in order to summarize the ADRs and rank them in order to determine the most relevant articles by the information provided in MEDLINE along with metadata corresponding to the articles. By retrieving articles about ADRs from PubMed, we are able to obtain recent scientific ADR summarization information about specific drugs. More detailed information for physicians can be provided when necessary by displaying links to the actual articles.
MeSH is a controlled vocabulary in MEDLINE, which indexes terms in biomedicine and health related areas19. The terms in MeSH are structured hierarchically. MeSH headings are concepts in the MeSH vocabulary, and many of them are used to index journal articles, and indicate the topics the articles discuss. MeSH subheadings are terms modifying and grouping MeSH headings, so that they can indicate a more specific sub-topic of the MeSH heading in an article. For example, ‘Rhabdomyolysis/Chemically Induced’ is a MeSH heading/subheading pair, and the subheading ‘chemically induced’ indicates the article discusses ‘Rhabdomyolysis’ specifically as an adverse effect.
Previous work has been done by our group to develop queries that retrieve articles related to ADRs for specific drugs20. Our method builds upon this work. In the previous study, a PubMed query is generated when given a drug name. The query includes the given drug name and pharmacological actions of the drugs combined with different key words, such as ‘adverse effects’ and ‘poisoning’. The query also includes filters to effectively rule out some false positive results. The retrieved articles achieved a precision of 93% and sensitivity of 90% in an initial evaluation.
The purpose of this work is to develop a method to extract information about ADRs and related MEDLINE reference to articles of given drugs, and present the most relevant information through a user-friendly interface in order to assist physicians in obtaining information about ADRs of drugs. Our method takes advantage of MEDLINE by retrieving related abstracts of articles, and by using MeSH headings and subheadings to extract ADRs from MEDLINE abstracts, as well as other information associated with the articles. The final display presents the information in an order of significance of the information about ADRs in order to facilitate the readability.
Method
An overview of this method is illustrated in Figure 1. For a given drug, we first use the query generator to retrieve all the articles related to the ADRs associated with it using the query described by Adams et al. 20. Then we use the ADR extractor to search through the PubMed summary of the article to extract information concerning the specified ADRs and other information associated with the articles which are needed for ranking. Within the MEDLINE citations of an abstract in PubMed, there are different sections that include information about the abstract. We recognize sections ‘JT’ (journal title), ‘DP’ (date of publication), ‘PT’ (publication type), and ‘MH’ (MeSH heading) to extract related information. In the third step, the ADRs and the articles mentioning them are ranked by the score given according to the information associated with the articles, such as publication date, number of citations, and influence factors. In the fourth step, the ADRs are displayed in a summary in order of the rankings and are grouped by body systems, which are determined using the MeSH tree.
Figure 1.
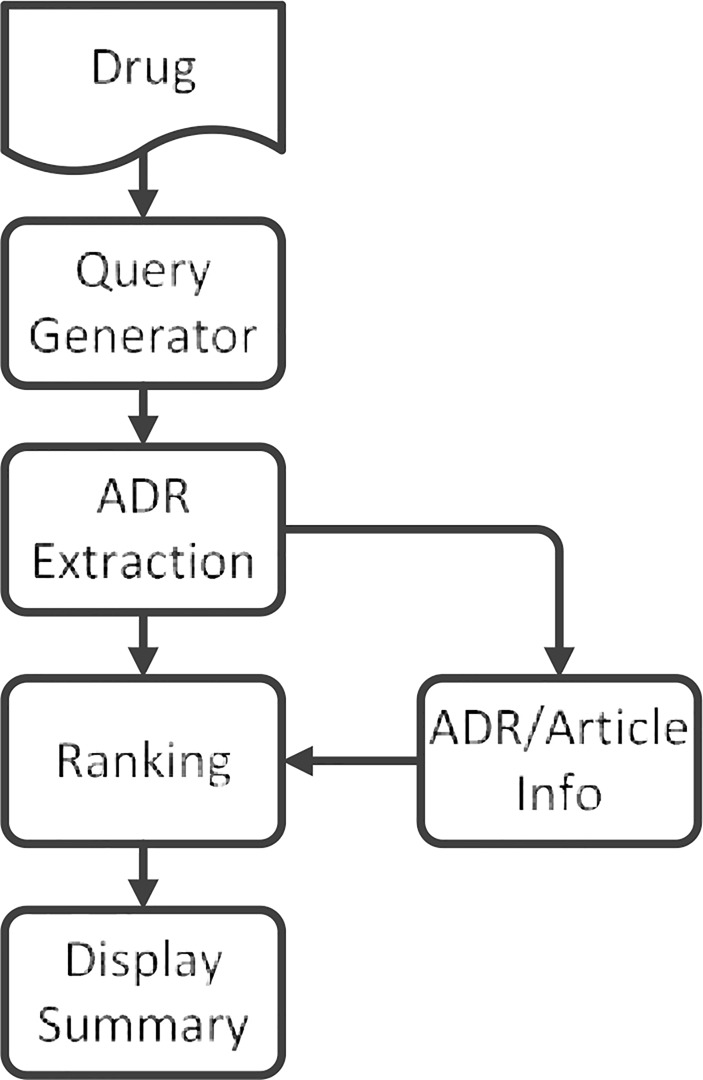
Method Overview
Query generator and article retrieval.
In the first step we use the query generator to generate a query as described by Adams et al. Given a drug, the query returns the PubMed identifiers (PMID) of the ADR articles for a specific drug that were retrieved from PubMed. Then we used E-utility, which is an application programming interface (API) (http://www.ncbi.nlm.nih.gov/books/NBK25500/) available in PubMed that generates URLs to extract content in PubMed to download the summaries of the articles from PubMed using their PMIDs.
ADR and information extraction.
In the second step, we extract information about ADRs from the PubMed abstracts. First, for each article, we determine the ADRs from the citations of abstracts in MEDLINE by using the MeSH heading/subheading pairs in the MEDLINE citations which begins with ‘MH’ (MeSH heading). An example is: ‘MH - Rhabdomyolysis/*chemically induced/*epidemiology’. Each MeSH heading is followed by a subheading which indicates the type of the MeSh heading. We determined that the MeSH headings with the subheading ‘chemically induced’ are ADRs. Those MeSH headings are assigned by the indexers because they represent biological phenomena or diseases induced by endogenous or exogenous substances. We also extract the MeSH headings beginning with ‘Drug-Induced’, such as ‘Drug-Induced Liver Injury’. In this step, we keep track of the articles, the corresponding ADRs, and the relevant information associated with them. The same ADRs can be mentioned in different articles, and we record the number of the times an ADR is mentioned and the articles that mention it.
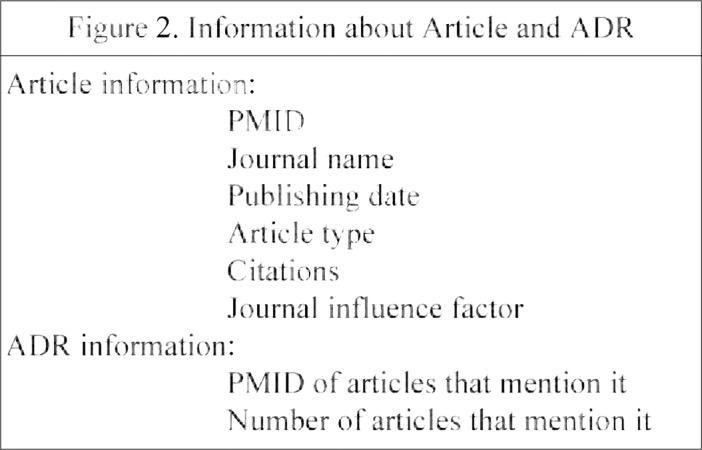
For the purpose of ranking and display, we extract different types of information for the articles, which are shown in Figure 2 and described below.
Figure 2.
Information about Article and ADR
PMID: obtained when the articles associated with the query are retrieved. For example, the PMID of an article mentioning the ADR rhabdomyolysis, which is associated with the medication Atorvastatin, is ‘15572716’.
Journal name: extracted from the MEDLINE summary of the article. The section containing it begins with ‘JT’, so that we extract the journal name by recognizing ‘JT - ‘ from the MEDLINE summary. For example, the journal title section of the article mentioned above is ‘JT - JAMA’.
Publishing date: also extracted from the MEDLINE summary of the article. The publishing date is in the section beginning with ‘DP’, so that we extract the publishing date by recognizing ‘DP -’ in the MEDLINE summary. For example, ‘DP - 2004 Dec 1’ is the publishing date section of the above article.
Article type: the type of the research article. It is described in the section beginning with ‘PT -’. For example, ‘PT - Journal Article’ is the type section of the article in the MEDLINE summary. Other types could be review articles or case reports.
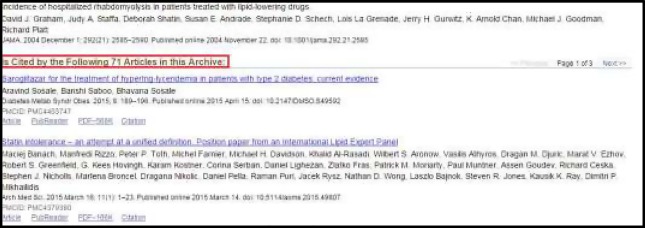
Citations: the number of times the article is cited by other articles in MEDLINE. The citations of an article are displayed in the PubMed page, so that we extract citations by reading the static html page (http://www.ncbi.nlm.nih.gov/pmc/articles/pmid/15572716/citedby/?tool=pubmed). Figure 3. illustrates the section that include citations in the PubMed page of the article.
Figure 3.
PubMed Page Indicating Citations
Influence factor: the influence factor of the journal where the article is published. We obtain the journal influence factor by Thomson Reuters (http://wokinfo.com/products_tools/analytical/jcr/). We downloaded the journal influence factor table for 2014 to determine the influence factor for the journal by matching the name of the journal. For example, the influence factor for JAMA is 30.387.
The information collected above is organized for each ADR, and consists of:
The articles that mention the ADR, since one ADR can be mentioned in several different articles. For example, for the ADR ‘Rhabdomyolysis’ which is an ADR of ‘Atorvastatin’, there are 3 articles that mention it, where the PMIDs of them are ‘15572716’, ‘23778904’ and ‘22419147’.
The number of different articles that mention the ADR. Therefore the value is ‘3’ for the ADR ‘Rhabdomyolysis’ that corresponds to Atorvastatin.
Ranking.
As in the next step, we rank each of the articles retrieved using the information obtained in the previous step, which are the number of citations and the journal influence factor. The score for each article is calculated by multiplying the number of times the article is cited and the influence factor of the journal where the article is published in.
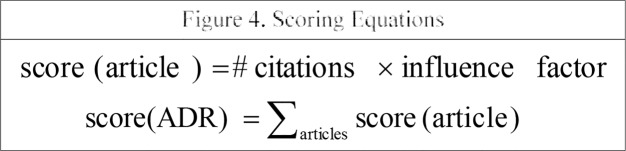
Last, we give each ADR a score it determine the significance of each ADR. The score of the ADR is given by the formula in Figure 4.
Figure 4.
Scoring Equations
For the ADR ‘Rhabdomyolysis’ mentioned above, the scores for each of the articles associated with it are: score(article 1) = 71 * 30.387 = 2157.477, score(article 2) = 7 * 16.104 = 112.728 and score(article 3) = 5 * 7.39 = 36.95. So the score of the ADR is: score(Rhabdomyolysis) = 2157.477 + 112.728 + 36.95 = 2305.155.
Display Summary.
The first step in displaying the summary involves grouping the ADRs by body systems. By grouping the ADRs of a drug, we are able to reduce the variety of ways of expressing similar ADRs in MeSH. For example, ‘Musculoskeletal Pain’ and ‘Muscular Diseases’ are both ADRs for the drug ‘Atorvastatin’ with similar meanings, and showing them both will be extra information for physicians to review. However, since they both belong to ‘Musculoskeletal Diseases’ body system, by displaying only the body system, we reduce the variety of similar ADRs.
To derive the body system of an ADR, we use the MeSH code corresponding to the ADR, which has been obtained from the MeSH tree. The MeSH codes can be divided into several parts and each part consists of three digits or characters, specifying a place in the MeSH hierarchy. The first three characters of the code represents the level of body system in the MeSH tree. Therefore we used the first three digits to determine the body system which we use to group the ADRs. For example, the MeSH code for ‘Rhabdomyolysis’ is ‘C10.597.617’, and ‘C10’ is the system level which stands for ‘Musculoskeletal Diseases’.
Some body systems in MeSH are divided into subclasses, so that to group further, we combine these subclasses. For example, ‘Female Urogenital Diseases’ and ‘Male Urogenital Diseases’ are combined into ‘Urogenital Diseases’. Some MeSH headings belong to several different body systems in MeSH, and in this situation, we choose the first body system.
Results
To evaluate performance of the system, we have tested our method on 6 drugs. The highest ranked ADR, and the articles associated with the ADR by ranking are summarized in Table 1. Articles are shown by their PMIDs. The article ‘15572716’, which discusses ‘Rhabdomyolysis’, received the highest score 2157 as a result of multiplying the journal influence factor 30.387 and the number of citations 71. The drugs and the number of ADRs found by the method are show in Table 2. The ADRs retrieved using our method are correct, as shown in Table 1. The method on average returned 4.8 ADRs for each drug for the 6 drugs we tested. At most 12 ADRs (‘Atorvastatin’) are found among the 6 drugs.
Table 1.
ADRs and articles for the 6 drugs
| Drug | ADR | Article | Score |
|---|---|---|---|
| Atorvastatin | Rhabdomyolysis | 15572716 | 2157 |
| 23778904 | 116 | ||
| 22419147 | 32 | ||
| Escitalopram | Arrhythmias Cardiac | 19556032 | 25 |
| Furosemide | Hypotension | 21035594 | 5 |
| Gabapentin | Exanthema | 17502552 | 132 |
| 18981374 | 33 | ||
| Metropolol | Atrioventricular Block | 18452693 | 23 |
Table 2.
Number of ADRs found for the 6 drugs
| Drug | Number of ADRs |
|---|---|
| Atorvastatin | 12 |
| Escitalopram | 4 |
| Furosemide | 3 |
| Gabapentin | 8 |
| Metropolol | 2 |
| Quinapril | 0 |
For the drug ‘Atorvastatin’, we obtained the results shown in Table 3. The score for each of the articles were obtained by multiplying the number of times the article was cited in PubMed and the journal influence factor. The score for each of the ADRs is the sum of the scores of the articles mentioning it.
Table 3.
ADRs and scores for Atorvastatin
| ADR | Article | Journal | Influence Factor | Article Score | ADR Score |
|---|---|---|---|---|---|
| Rhabdomyolysis | 15572716 | JAMA | 30.387 | 2157 | 2305 |
| 23778904 | Annals of internal medicine | 16.104 | 116 | ||
| 22419147 | Clinical pharmacology and therapeutics | 7.39 | 32 | ||
| Diabetes Mellitus Type 2 | 23704171 | BMJ | 16.378 | 376 | 744 |
| 21453832 | Journal of the American College of Cardiology | 15.343 | 368 | ||
| Acute Kidney Injury | 23511950 | BMJ | 16.378 | 393 | 505 |
| 23778904 | Annals of internal medicine | 16.104 | 112 | ||
| Musculoskeletal Pain | 23183941 | Circulation | 14.948 | 224 | 224 |
| Muscular Diseases | 16377285 | The American journal of cardiology | 3.425 | 47 | 47 |
| Albuminuria | 16377285 | The American journal of cardiology | 3.425 | 47 | 47 |
| Pain | 16377285 | The American journal of cardiology | 3.425 | 47 | 47 |
| Hematuria | 16377285 | The American journal of cardiology | 3.425 | 47 | 47 |
| Proteinuria | 15464670 | The American journal of cardiology | 3.425 | 27 | 27 |
| Diabetes Mellitus | 21257003 | The American journal of cardiology | 3.425 | 13 | 13 |
| Glucose Intolerance | 24152688 | The Journal of clinical endocrinology and metabolism | 6.31 | 12 | 12 |
| Hyperinsulinism | 24152688 | The Journal of clinical endocrinology and metabolism 30.387 | 6.31 | 12 | 12 |
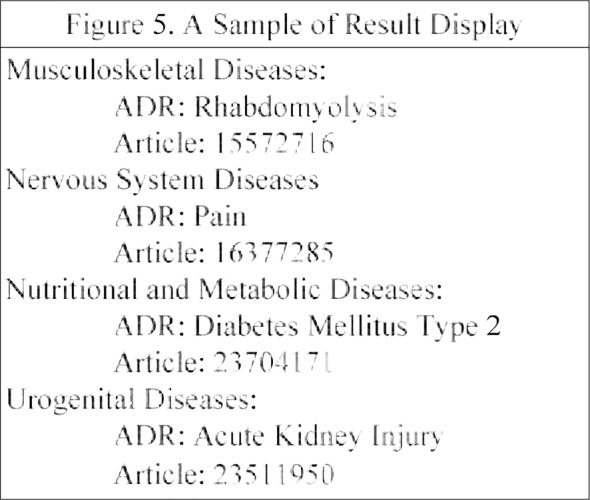
The body systems of ADRs are show in Table 4. The result is displayed hierarchically according to the body system, ADR and the corresponding article. For each body system, the most important ADR and corresponding article is displayed. A sample of the display format is illustrated below in Figure 5.
Table 4.
ADRs of Atorvastatin and body systems
| Drug | Body System | ADR |
|---|---|---|
| Atorvastatin | Musculoskeletal Diseases | Rhabdomyolysis |
| Musculoskeletal Pain | ||
| Muscular Diseases | ||
| Nervous System Diseases | Pain | |
| Nutritional and Metabolic Diseases | Diabetes Mellitus Type 2 | |
| Diabetes Mellitus | ||
| Glucose Intolerance | ||
| Hyperinsulinism | ||
| Urogenital Diseases | Acute Kidney Injury | |
| Proteinuria | ||
| Albuminuria | ||
| Hematuria | ||
| Escitalopram | Cardiovascular Diseases | Arrhythmias Cardiac |
| Eye Diseases | Glaucoma Angle-Closure | |
| Uveal Diseases | ||
| Nervous System Diseases | Seizures | |
| Furosemide | Cardiovascular Diseases | Hypotension |
| Pathological Conditions | Intraoperative Complications | |
| Skin and Connective Tissue Diseases | Sweet Syndrome | |
| Gabapentin | Skin and Connective Tissue Diseases | Exanthema |
| Stomatognathic Diseases | Stevens-Johnson Syndrome | |
| Behavior and Behavior Mechanisms | Self-Injurious Behavior | |
| Delusions | ||
| Mental Disorders | Depression | |
| Musculoskeletal Diseases | Fractures Bone | |
| Nervous System Diseases | Epilepsy Absence | |
| Status Epilepticus | ||
| Metropolol | Cardiovascular Diseases | Atrioventricular Block |
| Bradycardia |
Figure 5.
A Sample of Result Display
Discussion
In this work, we developed a method to extract and summarize ADRs from PubMed articles, and a ranking method to rank the ADRs as well as the articles to display the most relevant to physicians.
As shown in the results, the method successfully retrieved accurate ADRs for given drugs. For instance, for ‘Atorvastatin’, the method returned 12 ADRs. This shows the ability of this method to determine ADRs mentioned in the articles. In addition, the ranking aims to show the significance of the ADRs, and are meant to be helpful to physicians since they aim to be discriminative and informative. The scores include the information of both the number of articles mentioning an ADR and the importance of each article by summing the scores of all the articles. As shown in Table 2, generally speaking, the more articles mentioning an ADR, the more important the ADR may be. In addition, the higher quality of a journal, as determined by the journal’s impact factor, is important in determining the quality of the evidence associated with the ADR, and will be important to present to the physicians so that they will have more confidence in the evidence presented by the article. The body systems of ADRs indicate the body part where the ADR occurs and the body system that the drug most likely to have negative effects on. The grouping should be helpful to the physicians because it reduces the information they are presented, but this will have to be studied in future work.
There are some limitations in this study which are discussed below.
MeSH subheading.
We only use the MeSH subheading ‘chemically induced’ to identify the ADRs, and this may result in false negatives. Some articles retrieved by the query containing information about ADRs do not include MeSH headings modified with the subheading ‘chemically induced’, and therefore we cannot extract ADRs from them. This situation may be caused by the fact that some ADRs may not be a specific disease or symptom that is indexed in MeSH. For example, for drug ‘Quinapril’, no ADR was found by the method, but the article discussed adverse reactions, such as slowing recovery from postoperative anemia and increasing the incidence of cardiovascular events. Further work will be necessary to obtain these ADRs using textual features of the articles.
Body system.
When determining the body system where an ADR occurs in, we chose the first body system an ADR belonged to. In this situation, we lose some information about the body system of an ADR, and this may not be optimal. For example, ‘Pain’ which is an ADR of ‘Atorvastatin’, belongs to ‘Nervous System Diseases’, ‘Pathological Conditions, Signs and Symptoms’, ‘Psychological Phenomena and Processes’ and ‘Musculoskeletal and Neural Physiological Phenomena’. Our method recognized its body system as ‘Nervous System Diseases’, but the remaining choices also contain important information such as ‘Musculoskeletal and Neural Physiological Phenomena’.
Query dependency.
This method depends on the articles retrieved by the query, and performance of the query is critical. If some ADR articles are missed, the method will lose some ADR information. For example, 1 article was retrieved for ‘Quinapril’ and the method was unable to retrieve any ADR associated with it. Therefore, we may miss some ADRs due to the only dependency on the query.
Evaluation.
In the current stage, we have not formally evaluated the result of this method or experimented with aspects of the display. In future work, we will continue evaluation of the performance of the method with a larger set of drugs. In addition, further development followed by a usability evaluation in clinical practice will be needed to observe the utility and efficiency of the overall method and tool. The article scoring algorithm is based on heuristic algorithms and requires formal evaluation including identification of optimal input parameters.
The proposed approach may potentially have multiple applications. First, our methodology can be used to implement an interactive ADR dashboard providing most relevant publications related to a particular drug. A user interface for such a dashboard was previously described22. During recent years systematic access to verified drug information including ADRs has been significantly simplified by wide availability of various online and mobile tools23. However, for obvious reasons, these tools include only guideline-concordant information supported by expert consensus. Thus, emerging information about new ADRs of existing drugs or recent information about newly introduced drugs is usually not presented in these resources until fully verified. This kind of information can be accessed primarily via research publications23. The proposed interactive dashboard will be useful in this case because it will automatically perform literature search and present results in a summarized way sparing a clinician from necessity to spend time on running multiple PubMed queries and reviewing results. Second, current online compendia of drug information usually provide broad overviews without specific attention to particular subgroups such as children, older adults, minorities, or other groups. As such categories are easily identifiable using MeSH terms, the proposed dashboard can provide group specific searches addressing ADRs of a particular drug in a specific sub-group. Third, current electronic health records (EHR) increasingly include tools supporting knowledge delivery at point of care. Embedding the proposed dashboard as additional EHR functionality can help with delivering most recent evidence-based information about ADRs at the point of care. Fourth, there is growing number of initiatives aimed at aggregation of large number of EHRs from different health care systems such as the Observational Health Data Sciences and Informatics (OHDSI) collaborative (http://www.ohdsi.org/). These aggregated EHR repositories are used for knowledge discovery including ADRs. Semantic relationships including information about potential ADRs are essential components of automated knowledge discovery process. These repositories employ large knowledge bases which are automatically updated. The proposed approach may be used as a part of this process.
Significant further work is required in the future to fully explore the potential of the proposed approach including the following tasks: (1) evaluate usefulness of the proposed approach under different real-life scenarios (2) demonstrate higher performance than existing approaches such as just googling the drug name and the word ‘side effects’, (3) show that what is reported did not exist in current knowledge bases that are often used in EHR systems, or (4) perform a study of end users to assess their satisfaction and use of the system.
Conclusion
The study implemented a method to summarize the ADRs of a given drug, rank the retrieved articles and ADR information, and present the information concerning the ADRs in a simplified format. The information about ADRs was extracted from MEDLINE citations of journal articles using MeSH. The results demonstrated the feasibility and sufficient accuracy of this approach. The proposed approach can be used for automated summarization of ADR information from recent publications to facilitate translation of academic research into actionable information at point of care.
References
- 1.Naranjo C A, Busto U, Sellers E M, et al. A method for estimating the probability of adverse drug reactions. Clinical Pharmacology & Therapeutics. 1981;30(2):239–245. doi: 10.1038/clpt.1981.154. [DOI] [PubMed] [Google Scholar]
- 2.Shojania K G, Duncan B W, McDonald K M, et al. Safe but sound: patient safety meets evidence-based medicine. JAMA. 2002;288(4):508–513. doi: 10.1001/jama.288.4.508. [DOI] [PubMed] [Google Scholar]
- 3.To Err Is Human: Building a Safer Health System. National Academies Press; 2000. [PubMed] [Google Scholar]
- 4.Bates D W, Cullen D J, Laird N, et al. Incidence of adverse drug events and potential adverse drug events: implications for prevention. JAMA. 1995;274(1):29–34. [PubMed] [Google Scholar]
- 5.Gandhi T K, Weingart S N, Borus J, et al. Adverse drug events in ambulatory care. New England Journal of Medicine. 2003;348(16):1556–1564. doi: 10.1056/NEJMsa020703. [DOI] [PubMed] [Google Scholar]
- 6.Fattinger K, Roos M, Vergères P, et al. Epidemiology of drug exposure and adverse drug reactions in two Swiss departments of internal medicine. British journal of clinical pharmacology. 2000;49(2):158–167. doi: 10.1046/j.1365-2125.2000.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Classen D C, Pestotnik S L, Evans R S, et al. Computerized surveillance of adverse drug events in hospital patients. JAMA. 1991;266(20):2847–2851. [PubMed] [Google Scholar]
- 8.Einbinder JS, Bates DW. Leveraging information technology to improve quality and safety. IMIA Yearbook. 2007;2:22–29. [PubMed] [Google Scholar]
- 9.Hunt D L, Haynes R B, Hanna S E, et al. Effects of computer-based clinical decision support systems on physician performance and patient outcomes: a systematic review. JAMA. 1998;280(15):1339–1346. doi: 10.1001/jama.280.15.1339. [DOI] [PubMed] [Google Scholar]
- 10.McDonald CJ. Protocol-based computer reminders, the quality of care and the non-perfectability of man. The New England Journal of Medicine. 1976;295(24):1351–1355. doi: 10.1056/NEJM197612092952405. [DOI] [PubMed] [Google Scholar]
- 11.Wright A, Feblowitz J, Phansalkar S, et al. Preventability of adverse drug events involving multiple drugs using publicly available clinical decision support tools. American Journal of Health-System Pharmacy. 2012;69(3):221–227. doi: 10.2146/ajhp110084. [DOI] [PubMed] [Google Scholar]
- 12.Golder S, McIntosh H M, Loke Y. Identifying systematic reviews of the adverse effects of health care interventions. BMC medical research methodology. 2006;6(1):22. doi: 10.1186/1471-2288-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Haerian K, Salmasian H, et al. A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. AMIA Annu Symp Proc. 2011;2011:1464–70. [PMC free article] [PubMed] [Google Scholar]
- 14.Shetty KD, Dalal SR. Using information mining of the medical literature to improve drug safety. J Am Med Inform Assoc. 2011;18(5):668–74. doi: 10.1136/amiajnl-2011-000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurulingappa H, Rajput A M, Roberts A, et al. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. Journal of Biomedical informatics. 2012;45(5):885–892. doi: 10.1016/j.jbi.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Gurulingappa H, Mateen-Rajput A, Toldo L. Extraction of potential adverse drug events from medical case reports. J Biomed Semantics. 2012;3(1):15. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White KS, Lindsay A, Pryor TA, et al. Application of a computerized medical decision-making process to the problem of digoxin intoxication. Journal of the American College of Cardiology. 1984;4(3):571–576. doi: 10.1016/s0735-1097(84)80104-7. [DOI] [PubMed] [Google Scholar]
- 18.Katcher BS. MEDLINE: a guide to effective searching in PubMed and other interfaces. Ashbury Press; 2006. [Google Scholar]
- 19.Medical Subject Headings (MeSH) Fact Sheet. National Library of Medicine. https://www.nlm.nih.gov/pubs/factsheets/mesh.html.
- 20.Winnenburg R, Sorbello W, Ripple A, et al. Leveraging MEDLINE indexing for pharmacovigilance - Inherent limitations and mitigation strategies. JAMIA. 2015;57:425–435. doi: 10.1016/j.jbi.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams H, Friedman C, Finkelstein J. Automated Determination of Publications Related to Adverse Drug Reactions in PubMed. AMIA Jt Summits Transl Sci Proc. 2015;2015:31–5. [PMC free article] [PubMed] [Google Scholar]
- 22.Finkelstein J, Adams HZ, Chen Q, Lin K, Friedman C. Implementing automated delivery of evidence-based medication safety information to the point of care; Proceedings of the American Medical Informatics Association 2015 Annual Symposium; 2015. [Google Scholar]
- 23.Badgett RG, Dylla DP, Megison SD, Harmon EG. An experimental search strategy retrieves more precise results than PubMed and Google for questions about medical interventions. PeerJ. 2015;3:e913. doi: 10.7717/peerj.913. [DOI] [PMC free article] [PubMed] [Google Scholar]