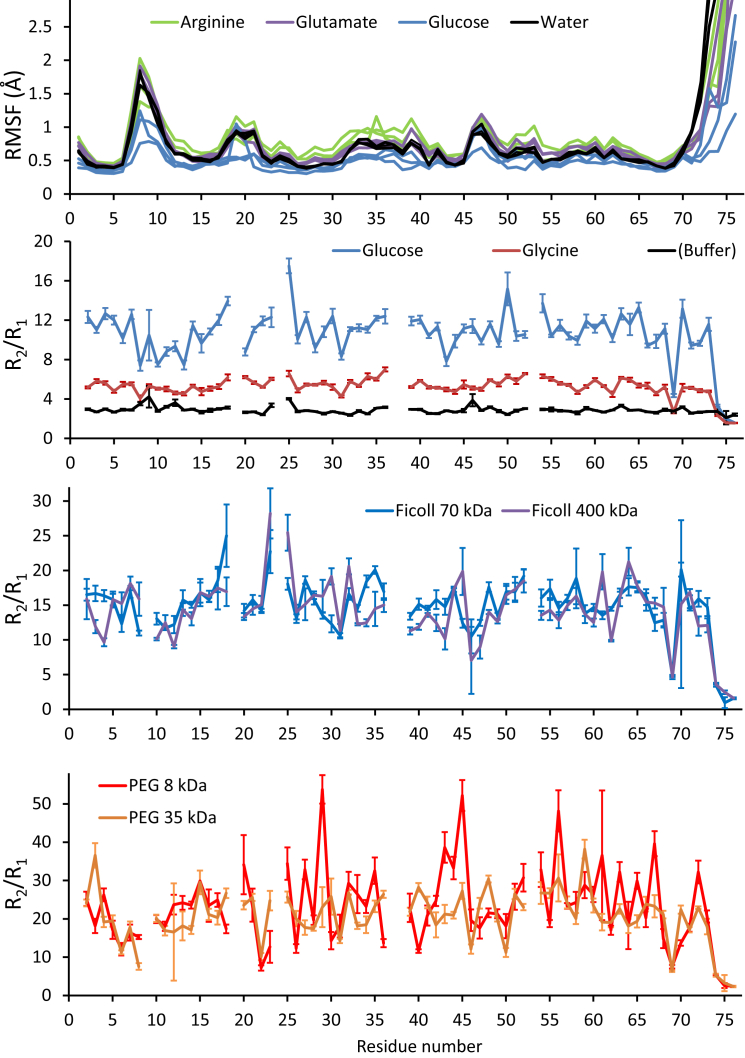
Figure 4.
Fast ubiquitin dynamics as observed in simulations (top; root mean-square fluctuation of Cα atoms) and by 15N relaxation (R2/R1 ratios). All systems contain 300 g/L of the indicated solute. In each R2/R1 profile, a positive deviation from the average indicates slow (microsecond- to millisecond-timescale) dynamics, whereas a ratio smaller than the average indicates fast (picosecond to nanosecond) fluctuations. Gaps correspond to weak or absent crosspeaks and three prolines in ubiquitin’s sequence; error bars correspond to one standard error propagated from T1 and T2 uncertainties. The median value for each R2/R1 profile reflects the global tumbling rate in those conditions (provided in Table S1 as the global correlation time, which is larger for slower diffusional rotation), which combines direct effects from viscosity and from dynamic association with other molecules in the solution, i.e., weak interactions. In the root mean-square fluctuation plots from MD simulations, higher values indicate sequence segments of high flexibility in the sampled timescale (picosecond to submicrosecond). To see this figure in color, go online.