Figure 1. Direct duplex detection methods join a host of powerful tools for RNA structure analysis.
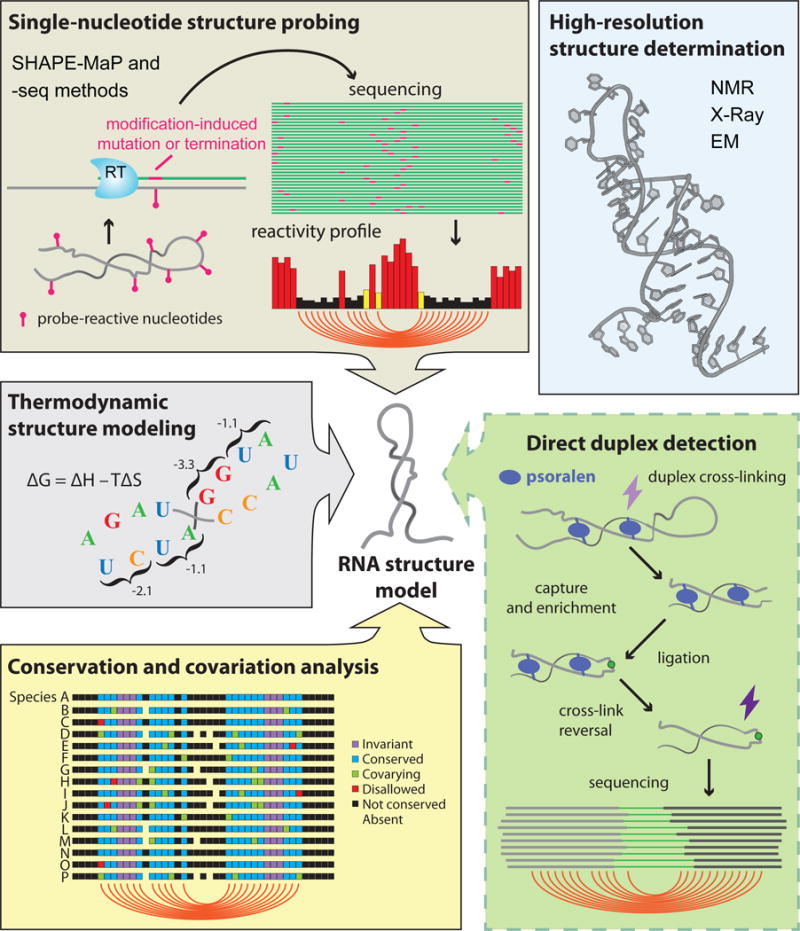
NMR, X-Ray crystallography, and cryo-EM can be used to determine atomic-resolution structures for “well-behaved” RNAs (blue box). For most other RNAs, a suite of complementary methods facilitates secondary structure determination. RNA structure can be probed with chemical or enzymatic reagents that, when read out by sequencing, provide a nucleotide-resolution profile of RNA flexibility (SHAPE-MaP and -seq methods, brown box). Thermodynamic stability parameters can be used to estimate the minimum energy structure for an RNA sequence (gray box). Base-paired nucleotides can be inferred from conservation patterns observed in sequence alignments of homologous RNAs (yellow box). This existing toolset is complemented by new direct duplex detection methods, in which paired RNA strands are cross-linked with psoralen and ligated together before detection via sequencing (green box).
