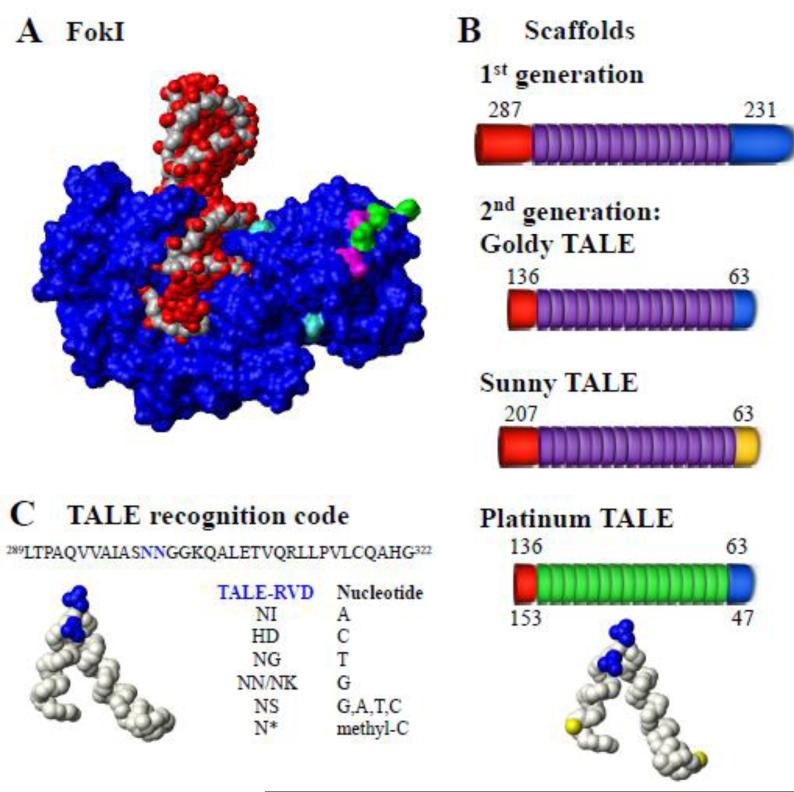
Fig. 3. Location of Sharkey and obligate heterodimer mutations in the FokI endonuclease.
A. Structure of complex (Protein Data Bank [PDB] accession = 1FOK) of wild type Flavobacterium okeanokoites FokI protein (Blue) with DNA (red/grey). Mutations are colored: magenta and green indicate the ELD and KKR obligate heterodimers mutations of the FokI, respectively [3]; Turquoise indicates the Sharkey mutations [3]. B. TALE recognition code. Protein sequence and structure of PthXo1 TALE (PDB accession = 3UGM, amino acids 289-322) with positions of the RVD colored blue(Mak et al. 2012). Nucleotide recognition code for substituted RVDs is shown; * indicated deletion. C. Scaffolds used to build TALENs. Lengths of N- and C-terminal domains are shown with TALE repeats. Red indicates the C-terminus; purple indicates 1st generation TALE repeats and green indicates Platinum TALE repeats; blue indicates 1st generation C-terminal domain; and yellow indicates mutated Sunny TALE C-terminal domain. PthXo1TALE structure (amino acids 289-322) with positions of the RVD (blue) and substitutions in the repeat scaffold at positions 4 and 35 (yellow) shown for Platinum TALEs (PDB accession = 3UGM). Structure figures were created with MOLMOL 2K.1 (Koradi et al. 1996).