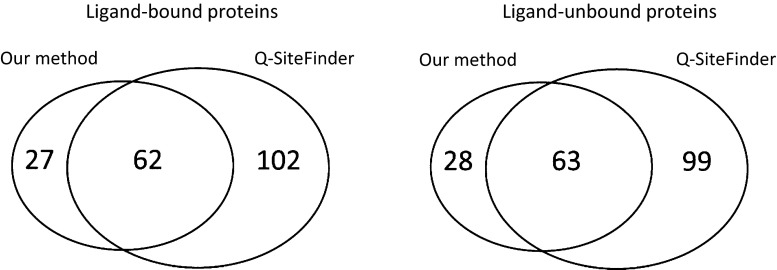
Fig. 5.
Numbers of proteins for which prediction by Q-SiteFinder and our proposed method failed. (Left) Prediction results for 342 ligand-bound structures (for ligand-binding space prediction). (Right) Prediction results for 348 ligand-unbound structures (for ligand-binding residue prediction). For both methods, the numbers of failed proteins are similar for ligand-bound and ligand-unbound proteins