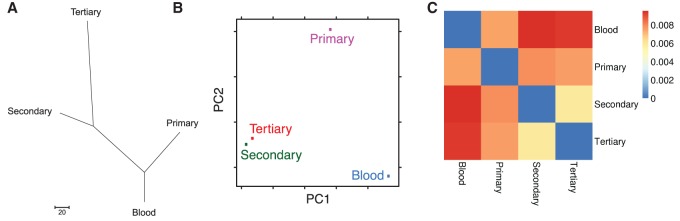
Figure 3.
Intersample relationships. (A) Phylogenetic tree. The Maximum Parsimony tree was obtained using the Max-mini branch-and-bound algorithm. The branch lengths were calculated using the average pathway method. Evolutionary analyses were conducted in MEGA6. (B) Principal coordinate analysis. The x- and y-axes represent the first and second coordinates, respectively. In this case, the first and second principal coordinates are genetic variation and somatic mutation, respectively. (C) Heatmap displaying total root-mean-square deviation of each sample's variant allele frequencies from each other.