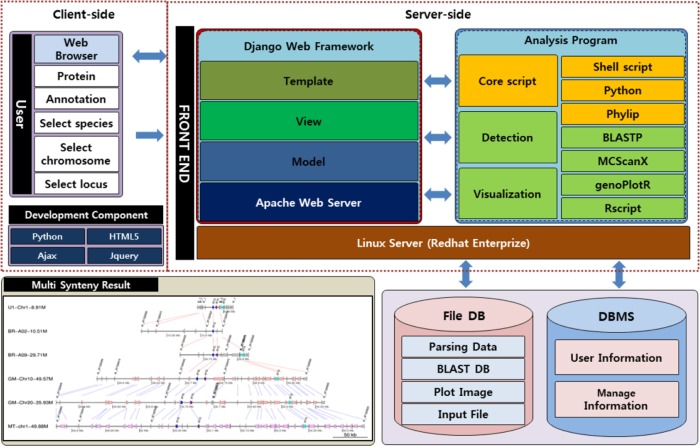
Figure 1.
Architecture of MultiSyn webtool.
Notes: Architecture shows two configurations, the client-side and server-side. The client-side provides the interface for input and output for the MultiSyn user using HTML5 and jQuery. The user performs an analysis to enter the proteins (pep) and annotation (simplified BED) file or select species, chromosome, locus in a Web browser. The server-side consists of the core script for processing in connection with the webtool and for the determination and visualization of synteny. In the core script using python, phylip, shell script to connect the user to enter data and other utilities to detect and visualize synteny treated as a result. Input information is stored in the user information for the file DB and the analysis conducted provides the final output product.