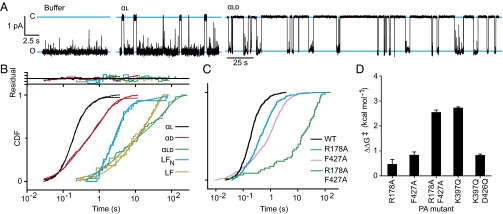
Fig. 3.
Peptide stereochemistry and channel mutational analysis of translocation. (A) Single-channel current records (black line) under an asymmetric ΔpH gradient of 2 units (pHcis 5.6 and pHtrans 7.6) and an asymmetric KCl gradient (100 mM [added KCl]cis, 0 mM [added KCl]trans) for (Left) buffer alone, (Middle) 20 nM αl, and (Right) 60 nM αld peptides. Open (O) and closed (C) states are designated with blue lines. Note that scale bar for buffer and αl is given at left; larger time scale bar is given for αld, but the current scale is the same. (B) CDF of the single-channel blocked-state dwell times for the following peptide substrate and channel combinations: WT PA and αl (black), αd (red), αld (green), LFN (blue), and LF (orange). Residuals to chemical kinetic modeling fits are given at top. (C) CDF of the single-channel of the blocked-state dwell times for αl peptide via WT (black) and the following mutant channels: PA R178A (blue), PA F427A (magenta), and PA R178A F427A (green). Individual CDFs were reproduced two to five times on individual membranes. (D) Activation free energy differences [i.e., ΔΔG‡ = ΔG‡(mutant PA) - ΔG‡(WT)] for PA channel mutants from WT for the translocation of αl peptide. ΔΔG‡ errors are propagated from SDs of t1/2 values based on three to five CDF measurements per mutant.