Significance
The yeast HO gene is under complex regulation during the cell cycle. Swi5 binds to the promoter during the M phase to promote later activation but is absent when the gene is activated in late G1. Thus cells “remember” the effects of Swi5 at the promoter, and this is especially important during prolonged G1 arrest due to nutrient deprivation. Promoter “memory” consists of SBF factor and Mediator bound at the promoter. However, this promoter memory is lost in cells arrested in G1 by pheromone exposure. We show that this memory loss occurs by transcription through the promoter, displacing bound SBF, thereby erasing the memory. Thus, factors at a promoter can constitute memory of prior decisions, and displacement by transcription can remove this memory.
Keywords: lncRNA, nucleosomes, promoter memory, gene regulation, chromatin
Abstract
The yeast HO endonuclease is expressed in late G1 in haploid mother cells to initiate mating-type interconversion. Cells can be arrested in G1 by nutrient deprivation or by pheromone exposure, but cells that resume cycling after nutrient deprivation or cyclin-dependent kinase (CDK) inactivation express HO in the first cell cycle, whereas HO is not expressed until the second cycle after release from pheromone arrest. Here, we show that transcription of a long noncoding RNA (lncRNA) mediates this differential response. The SBF and Mediator factors remain bound to the inactive promoter during arrest due to CDK inactivation, and these bound factors allow the cell to remember a transcriptional decision made before arrest. If the presence of mating pheromone indicates that this decision is no longer appropriate, a lncRNA originating at –2700 upstream of the HO gene is induced, and the transcription machinery displaces promoter-bound SBF, preventing HO transcription in the subsequent cell cycle. Further, we find that the displaced SBF is blocked from rebinding due to incorporation of its recognition sites within nucleosomes. Expressing the pHO-lncRNA in trans is ineffective, indicating that transcription in cis is required. Factor displacement during lncRNA transcription could be a general mechanism for regulating memory of previous events at promoters.
The yeast HO gene encodes an endonuclease that initiates interconversion of the mating type (MAT) locus by recombinational gene conversion (1). Inappropriate expression of an endonuclease can damage DNA, so the HO gene is under extremely tight repression, requiring multiple activators and coactivators for activation of transcription (2). The regulatory region of the HO gene is exceptionally large and complex by yeast standards (Fig. 1A). Most yeast genes have promoter regions of 300–500 bp, whereas at the HO locus the closest gene is nearly 3 kb away. Chromatin plays a critical role in HO regulation, with activation requiring waves of nucleosome eviction along the promoter during the cell cycle (3, 4).
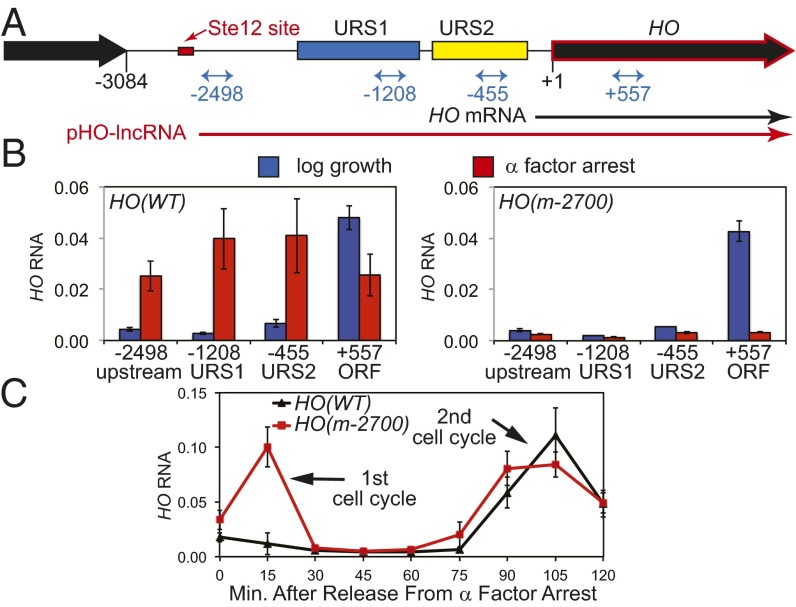
Fig. 1.
An Ste12 binding site is required for a lncRNA at HO. (A) Map of the HO promoter with the Ste12 binding site at –2700 indicated in red and locations of the RT-qPCR primers indicated in blue. The HO mRNA is black, and the pHO-lncRNA is red. (B) RNA measurements at locations in the HO promoter and ORF in HO(WT) and HO(m-2700). Blue bars indicate log growth, and red bars indicate α-factor arrest. (C) HO(WT) (black) and HO(m-2700) (red) strains were released from α-factor arrest and HO RNA measured as a function of minutes after release.
The yeast HO gene shows a complex pattern of transcriptional regulation (for review, see ref. 2). First, HO transcription is cell cycle-regulated, with expression limited to a short period in late G1. This regulation follows function, as mating-type interconversion occurs in G1 phase, before the MAT locus is replicated during S phase. Cleavage of one copy of MAT after S phase would favor homologous recombination with the sister chromatid over the desired gene conversion event and could cause other problematic events. Second, HO is only expressed in haploid cells; mating-type interconversion is exclusive to haploids as a means to facilitate mating and diploid formation. Finally, HO is expressed only in mother cells but not in daughters. This asymmetry in mating-type interconversion, along with axial budding that occurs only in haploid cells, ensures that two mitotic divisions of a spore will produce four cells, two MATa and two MATα, positioned to promote mating and efficient diploidization (5). Thus, regulation of HO is tightly governed to prevent endonuclease expression, except in specific cells for a limited time during the cell cycle.
ChIP experiments have shown a complex series of events at the HO promoter during cell-cycle progression (for review, see ref. 2). The Swi5 DNA-binding transcription factor initiates the cascade of HO activation, entering the nucleus as cells progress through anaphase. Swi5 binds to sites at –1816 and –1305 within the URS1 region of the HO promoter, where it recruits three transcriptional coactivators, SAGA, SWI/SNF, and Mediator, leading to nucleosome loss at URS1. Swi5 is then quickly degraded, and the cessation of Swi5 binding is accompanied by loss of coactivator recruitment at HO URS1. The coactivators that had been recruited to URS1 trigger subsequent “waves” of nucleosome loss, first in the upstream part of URS2 and then moving downstream (3). Nucleosome eviction in URS2 depends on the Swi4/Swi6 cell-cycle box binding factor (SBF) DNA-binding complex and two histone chaperones that facilitate SBF binding at URS2 (3, 4). Finally, SBF recruits multiple factors including SWI/SNF, SAGA and Mediator to promote the assembly of the transcription complex at TATA.
The Swi5 transcription factor binds transiently to the HO promoter during M phase. However, HO is only expressed late in the following G1 phase, after cells transit the START point of the cell cycle. Despite this pivotal role for Swi5 in promoting HO expression, there is no Swi5 protein bound to the HO promoter at the time the gene is transcribed. Thus, cells must “remember” that Swi5 was present earlier in the cell cycle, and experiments showed that this memory at the HO promoter can be long lasting (6).
Although the time between Swi5 degradation and HO expression is normally less than 20 min in rapidly growing cultures, it can be extended to many hours and even days. There are several ways to arrest yeast cells in G1 phase, including starvation and pheromone treatment. However, the ability of cells to express HO upon release from arrest depends on the manner in which the cells were arrested. Nutrient deprivation results in stationary phase cells that are arrested in G1, and these cells express HO when they eventually reenter the mitotic cell cycle when given fresh nutrients (7). These G1-arrested cells had progressed part way through the sequence of promoter events; Swi5 had already bound to the promoter, recruited coactivators, and nucleosomes were evicted from URS1 and URS2. We used ChIP assays to determine which proteins are bound to the HO promoter during a similar arrest caused by cyclin-dependent kinase (CDK) inactivation (8) and found that SBF and the Mediator complex are bound to URS2 during the arrest. Importantly, SWI/SNF and SAGA are not stably bound to URS2 during the arrest, and neither Swi5 nor coactivators were present at URS1. Thus, SBF and Mediator remain bound at the promoter, serving as “memory” in G1-arrested cells and allowing the promoter to be effectively expressed when cells reenter the cell cycle.
Cells can also be arrested in G1 by exposure to a mating pheromone such as α-factor, and cells synchronously reenter the cell cycle when mating pheromone is removed. Mating pheromone exposure activates a MAP kinase cascade resulting in phosphorylation of the Ste12 transcription factor and activation of Ste12 target genes (9, 10). However, there is a major difference in HO expression compared with release from other types of arrest: HO is not expressed in the first cell cycle following release from mating pheromone arrest, but HO is expressed in subsequent cycles (11).
This regulation of HO transcription is sensible from the teleological standpoint that diploidy is favored over haploidy. After a haploid cell has successfully mated to form a zygote, it will pass START and enter the cell cycle; expression of the HO endonuclease would wreak recombinational havoc in a diploid cell, and thus, a mechanism to prevent HO expression after release from mating pheromone arrest is sensible. This is supported by the observation that zygotes newly formed by conjugation between haploids are incapable of switching mating type during their first cell cycle following karyogamy (12). Similarly, if a haploid cell arrested by mating pheromone fails to mate, there are mechanisms that allow the cell to terminate the arrest and reenter the cell cycle (13). Under these circumstances, it makes sense for the cell to prevent HO expression and mating-type interconversion, as the cell has a potential mating partner in the vicinity, and it may successfully mate in the next G1 phase. Thus, for mating attempts that are either successful or unsuccessful, it is reasonable for the cell to block HO expression when reentering the cell cycle following mating pheromone arrest. Haploid cells therefore need to remember the decision to switch mating type when resuming growth after starvation-induced arrest but forget this decision when resuming growth after mating pheromone-induced arrest.
Long noncoding RNAs (lncRNAs) can regulate transcription (14–17), and in budding yeast lncRNAs have been implicated in promoter regulation (18–24). Here we decipher the mechanism by which mating pheromone blocks expression of the HO endonuclease during the subsequent cell cycle. We show that mating pheromone induces expression of a lncRNA initiated upstream of the promoter and that this transcription leads to loss of the SBF transcription factor bound at the promoter as well as increased nucleosome deposition at URS2. The activity of this lncRNA transcription could be an example of a more general mechanism for regulating “promoter memory” in other systems of complex temporal gene regulation.
Materials and Methods
All yeast strains used in this study are listed in Table S1 and are isogenic in the W303 background (25). To mutate the Ste12 binding site at –2700 from TGAAACA to CTCGAGG and to insert a 196-bp ADH1 terminator fragment at –2200, we used the delitto perfetto method (26). For α-factor arrest experiments, cells were grown in YM-1 medium (27) at 25 °C to early log phase, and α-factor (4.5–6 µM, depending on the batch) was added. Cells were monitored by light microscopy to determine when they were arrested, typically for 2–2.5 h, as determined by microscopy. After arrest, cells were filtered, washed with one volume of prewarmed YM-1 medium, and released into fresh YM-1 medium containing 0.15 mg/mL Pronase (Sigma, 81748) at 25 °C. For other experiments, cells were grown at 30 °C in YPAD medium [1% yeast extract, 2% (wt/vol) bactopeptone, 0.002% adenine, 2% (wt/vol) dextrose] (28).
Table S1.
Strain list
| Yeast strain | Genotype |
| Fig. 1 | |
| DY13725 | MATa ho::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY17320 | MATa ho(m-2700)::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| Fig. 2 | |
| DY13725 | MATa ho::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY17405 | MATa SWI4-V5::His3MX ho::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY17403 | MATa SWI4-V5::His3MX ho(m-2700)::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| Fig. 3 | |
| DY17405 | MATa SWI4-V5::His3MX ho::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY17922 | MATa SWI4-V5::His3MX ho(tADH1-2200)::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY17403 | MATa SWI4-V5::His3MX ho(m-2700)::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| Fig. 4 | |
| DY18370 | MATa/MAT∆::LEU2 HO/HO(m-2700;pFA6+450)::KanMX ade2 can1 his3 leu2 +/lys2 +/met15 trp1 ura3 |
| DY18371 | MATa/MAT∆::LEU2 HO(m-2700)::KanMX/HO(pFA6+450)::KanMX ade2 can1 his3 leu2 +/lys2 +/met15 trp1 ura3 |
| Fig. 5 | |
| DY17405 | MATa SWI4-V5::His3MX ho::KanMX ade2 can1 his3 leu2 trp1 ura3 |
| DY17532 | MATa SWI4-V5::His3MX CLN2[-764 to -435 deleted]:CLN1[-850 to -450] ho[-953 to -624 deleted]:CLN2[-764 to -435, wtNDR]::KanMX ade2 can1 his3 leu2 trp1 ura3 |
| DY17535 | MATa SWI4-V5::His3MX CLN2[-764 to -435 deleted]:CLN1[-850 to -450] ho[-953 to -624 deleted]:CLN2[-764 to -435, mutNDR]::KanMX ade2 can1 his3 leu2 trp1 ura3 |
| DY17403 | MATa SWI4-V5::His3MX ho(m-2700)::KanMX[3′] ade2 can1 his3 leu2 trp1 ura3 |
| DY18801 | MATa SWI4-V5::His3MX CLN2[-764 to -435 deleted]:CLN1[-850 to -450] ho(m-2700):[-953 to -624 deleted]:CLN2[-764 to -435, wtNDR]::KanMX ade2 can1 his3 leu2 trp1 ura3 |
| DY18802 | MATa SWI4-V5::His3MX CLN2[-764 to -435 deleted]:CLN1[-850 to -450] ho(m-2700):[-953 to -624 deleted]:CLN2[-764 to -435, mutNDR]::KanMX ade2 can1 his3 leu2 trp1 ura3 |
The promoter constructs in DY17532, DY17535, DY18801, and DY18802 have been described (30). The KanMX marker is inserted beyond the HO 3′ UTR.
RNA was measured by RT-quantitative PCR (RT-qPCR) as described (29) using random primed cDNA synthesis, with RNA expression normalized to RPR1 expression. ChIP and real-time qPCR were performed as described (29, 30). The Swi4-V5 ChIP values in Figs. 2 and 3 were first normalized to ChIPs at the CLN2 promoter, and then both types of ChIPs were normalized to their respective input DNA sample (30). All qPCR experiments were run on a Roche Lightcycler 480, and concentrations were determined using genomic DNA or ChIP input for in-run standard curves via the E-Method (31). Error bars in the RT-qPCR and ChIP assays reflect the SD of three biological samples. RT-qPCR and ChIP primers used in this study are listed in Table S2.
Fig. 2.
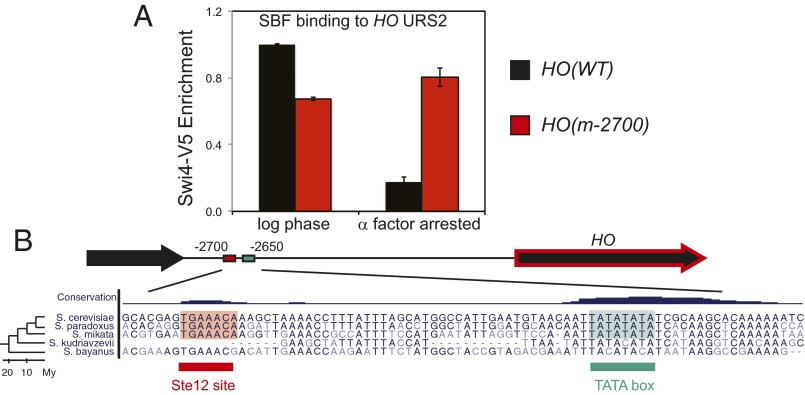
Mutating the Ste12 binding site allows SBF to bind to HO during arrest. (A) Swi4-V5 binding to HO URS2 was measured by ChIP in log phase and in α-factor–arrested cells for HO(WT) (black) and HO(m-2700) (red). (B) Conservation of HO lncRNA. A multiple alignment of the Ste12 binding site and TATA box ∼2,700 bp 5′ of HO. The whole genome alignments and conservation tracks were obtained from the UCSC genome browser. On the left, a phylogenetic tree shows the evolutionary distance between species.
Fig. 3.
A transcriptional terminator blocks the effect of the STE12-dependent lncRNA. (A) Map of the HO promoter with the position of the ADH1 terminator inserted at –2200 indicated in green and the locations of the RT-qPCR primers indicated in blue. (B) RNA measurements at locations in the HO promoter and ORF in HO(WT) and HO(tADH1-2200). Blue bars indicate log growth, and red bars indicate α-factor arrest. (C) Swi4-V5 binding to HO URS2 was measured by ChIP in log phase and in α-factor–arrested cells for HO(WT) and HO(tADH1-2200). Blue bars indicate log growth, and red bars indicate α-factor arrest. (D) HO(WT) (black) and HO(tADH1-2200) (red) strains were released from α-factor arrest, and HO RNA was measured as a function of time after release.
Table S2.
Oligonucleotide list
| Oligo | Description | Sequence |
| RT-qPCR oligos | ||
| F3307 | HO Upstream (–2545 to –2522) | CTATAAGGTTGATATTCTCACGAG |
| F3312 | HO Upstream (–2478 to –2447) | TAGCAATTATAATCTCTAATATACTATTTCTG |
| F1093 | HO URS1 (–1429 to –1410) | TATACCCAATCGCTGCGTGC |
| F1094 | HO URS1 (–1158 to –1139) | AGCCGCCACGAATCAAACTT |
| F1095 | HO URS2 (–825 to –806) | GGCAAACCTAATGTGACCGT |
| F1096 | HO URS2 (–508 to –489) | ACAGGACTTGCGAACCCTTT |
| F1066 | HO ORF (+452 to +474) | AAATGGAGCGCTCTAAAGGAGAA |
| F1067 | HO ORF (+639 to +661) | CTAACCACAGACCAAGCATCCAA |
| F3550 | HO(+pFA6 inserted) | GCATTAATGAATCGGCCAAC |
| F3551 | HO(+pFA6 inserted) | CAAGGTCACTACGTCCAGTGAG |
| F2430 | RPR1 control | CACCTATGGGCGGGTTATCAG |
| F2431 | RPR1 control | CCTAGGCCGAACTCCGTGA |
| ChIP-qPCR oligos | ||
| F1095 | HO URS2 | GGCAAACCTAATGTGACCGT |
| F1096 | HO URS2 | ACAGGACTTGCGAACCCTTT |
| F1959 | HO(-927) | GAGGTTGGTATTGATTGTTG |
| F1982 | HO(-828) | GTCAGGGTATGAACCATACG |
| F3235 | CLN2 | GTATCCTCCGCACTTTTACCC |
| F3236 | CLN2 | GCGCATGAATTGATAACTTGG |
| F1399 | IGR-V control | GGCTGTCAGAATATGGGGCCGTAGTA |
| F1400 | IGR-V control | CACCCCGAAGCTGCTTTCACAATAC |
The evolutionary conservation of binding sites was assessed by analyzing a whole genome alignment across seven yeast species, produced by the UCSC Genome Browser (32). The conservation scores are based on a Hidden Markov Model (phylo-HMM) (32).
Results
A lncRNA at the HO Promoter Regulates Binding of SBF.
In addition to cell-cycle regulation of native HO mRNA, two studies identified an RNA originating upstream of the normal HO transcription start site (33, 34). This upstream RNA is only detected in cells arrested by α-factor pheromone, suggesting it could provide a mechanism for the inhibition of HO expression following α-factor exposure. Because this RNA is absent from polysomes, it is considered to be noncoding (34).
Pheromone exposure leads to phosphorylation of the Ste12 transcription factor, which binds to promoters and induces expression of target genes (9, 10). The lncRNA expressed at HO is not present in an ste12 mutant (34), making Ste12 a good candidate for inducing this lncRNA. Consistent with this prediction, we identified an Ste12 binding site (TGAAACA) ∼2,700 bp from the ATG codon in the upstream region of the HO locus, which extends 3 kb to the nearest adjacent gene (Fig. 1A). There is a TATA element (TATATATA) about 50 bp downstream from the Ste12 site. We isolated RNA from α-factor arrested cells and used RT-qPCR with primers throughout the HO promoter. These results, along with northern data (33, 34), show this pHO-lncRNA originates upstream of our most upstream primer at –2547 and runs through the entire HO promoter (Fig. 1B). We mutated the Ste12 binding site by site-directed mutagenesis and inserted the resulting HO(m-2700) version in place of the natural promoter. RT-qPCR measurements show that α-factor does not induce expression of the HO noncoding RNA from the HO(m-2700) promoter (Fig. 1B). We conclude that the Ste12 binding site is required for pHO-lncRNA expression.
To determine whether this pHO-lncRNA regulates HO expression after release from pheromone arrest, we next performed α-factor arrest and release experiments with wild-type and the HO(m-2700) mutant promoters (Fig. 1C). In wild-type cells, HO is expressed in the second cycle following release, but not in the first cycle, consistent with a previous report (35). In contrast, HO is expressed in both cell cycles in the strain with the HO(m-2700) mutant promoter, which lacks the Ste12 binding site. This result suggests that pHO-lncRNA is required to prevent HO mRNA expression in the first cell cycle following release from pheromone arrest.
Nutrient deprivation or conditional CDK inactivation also causes G1 arrest, but this arrest is physiologically different from that caused by α-factor exposure. The ability to express HO after G1 arrest caused by CDK inactivation requires promoter memory, and as described in the introduction, the Swi5 activator that binds at URS1 is necessary for cells to build memory and to activate HO following release. We previously investigated the molecular nature of this memory and demonstrated that SBF and Mediator are stably bound to URS2 during this type of G1 arrest (8). We therefore determined whether SBF is bound to URS2 at the HO promoter during the G1 arrest induced by α-factor. SBF binds to both HO(WT) and HO(m-2700) in log-phase cells, but during α-factor arrest, SBF binds only to the HO(m-2700) promoter and not to the wild-type promoter (Fig. 2A). We conclude that the Ste12-induced expression of the pHO-lncRNA leads to loss of SBF binding at URS2.
Given the potentially crucial role for pHO-lncRNA in regulating HO expression, we investigated whether the pHO-lncRNA is evolutionarily conserved. We asked whether the Ste12 binding site and the TATA box upstream of HO were evident in other yeast species. Using a multiple-genome alignment across seven yeast species, we found that both the Ste12 and TATA motifs were exactly conserved in Saccharomyces cerevisiae, Saccharomyces paradoxus, and Saccharomyces mikatae, but not in Saccharomyces kudriavzevii and Saccharomyces bayanus (Fig. 2B). These results indicate conservation among close relatives of S. cerevisiae but not among more distant yeasts, which may have evolved independent mechanisms of HO regulation. This model is consistent with recent observations that the Ste12-mediated regulatory networks have been subject to high levels of turnover in fungi (36).
These results are consistent with a model in which the act of transcription of the pHO-lncRNA by RNA polymerase II (pol II) leads to displacement of SBF bound at the promoter. If this is true, then terminating transcription of the lncRNA upstream of the SBF binding sites should prevent SBF displacement by pol II. To directly test this model, we inserted an ADH1 terminator at –2200, upstream of URS1, creating HO(tADH-2200) (Fig. 3A). RT-qPCR analysis shows that the terminator does not affect HO expression in log phase [Fig. 3B, blue bars, compare HO(WT) to HO(tADH-2200)]. During α-factor arrest (Fig. 3B, red bars), RNA complementary to URS1 and URS2 is absent in the HO(tADH-2200) strain, whereas the RNA upstream of the terminator is still present. ChIP experiments show that SBF still binds to the HO(tADH-2200) promoter during α-factor arrest, whereas SBF is not bound at HO(WT) (Fig. 3C). Consistent with the proposed model, we conclude that pol II transcription through URS2 leads to loss of SBF binding.
We next asked whether promoter changes affected HO expression following release from α-factor arrest. Strains were arrested with α-factor and then released, and HO mRNA was measured during the time course (Fig. 3D). The wild-type promoter did not express HO following release, as expected, but in contrast, the HO(tADH-2200) promoter with a terminator inserted upstream of URS1 expressed abundant HO mRNA following release from arrest, as did the HO(m-2700) promoter with disrupted Ste12 binding site (Fig. 1C). Thus, there is a good correlation between SBF binding during α-factor arrest and HO expression following release.
Transcription of the pHO-lncRNA Acts in Cis to Regulate HO Expression.
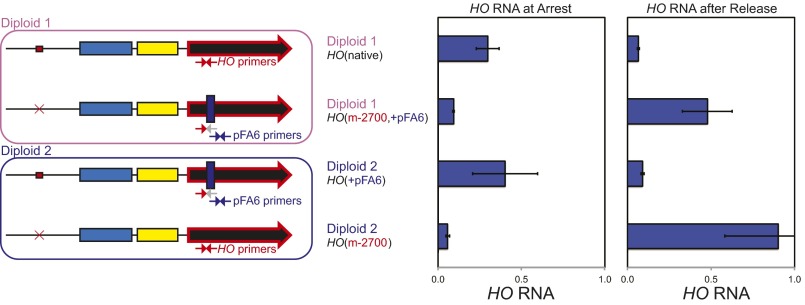
Given a potential link between pHO-lncRNA expression and SBF binding, we predict that pol II transcribing though URS2 during α-factor arrest directly displaces SBF, leading to a loss of promoter activity following release from pheromone arrest. Alternatively, the Ste12-induced lncRNA itself could act to displace SBF from binding, given that lncRNAs have been shown to act in trans to alter activity of enhancers in mammalian cells (16), and in yeast, at least one lncRNA can act in trans (20). We developed a cis/trans test to directly address this question for the pHO-lncRNA. We used diploid strains with two HO promoters, one native and one with the mutated STE12 binding site at –2700 (Fig. 4). To distinguish between these two alleles by RT-qPCR, we deleted the genomic region containing one of the primers within the ORF used to measure HO mRNA, and this region was replaced with a segment of plasmid pFA6 (37). Thus, we can use distinct primers to differentiate between the native HO allele and the allele with the pFA6 insert. Control experiments in haploids confirmed the specificity of the primers. To control for the possibility that the pFA6 insertion affected promoter activity, we constructed two diploid strains differing in which allele had the pFA6 insertion (Fig. 4). HO expression is strongly repressed in diploids by the a1/α2 repressor encoded by MATa and MATα (38, 39). To overcome this obstacle, we used MATa/mat∆ diploid strains that do not express a1/α2. RNA was isolated from these diploid strains arrested with α-factor and also at 15 min after release from arrest. RNA complementary to the HO ORF was detected from the native HO promoter at arrest but not following release (Fig. 4). In contrast, the opposite pattern was observed with the promoter lacking the STE12 binding site. Thus, the pHO-lncRNA induced at arrest from the native promoter did not affect activity after release of the HO(m-2700) promoter in the same cell. We conclude that transcription of the HO lncRNA functions in cis, and this result is consistent with transcription displacing promoter-bound transcription factors.
Fig. 4.
Transcription of the HO lncRNA acts in cis. (Left) A diagram of the two diploid strains and the HO alleles are shown. The alleles labeled HO(m-2700) have a mutation at the Ste12 binding site, and the alleles labeled HO(+pFA6) have a deletion within the HO ORF that removes the sequence for one of the HO RT-qPCR primers (indicated by the gray arrow) with this sequence replaced with the sequence from plasmid pFA6. RNA was isolated from the two diploid strains arrested with α-factor, and from cells 15 min after release, and quantitated by RT-qPCR. Distinct primer sets distinguish between the two alleles. This experiment required the use of a distinct cDNA standard prepared from these strains, due to the pFA6 primer sequences not being present in our standard genomic DNA prep, and this different DNA standard explains the higher RNA values compared with other experiments.
Promoter Nucleosomes Are Essential to Prevent HO Expression After Release.
Nucleosomes could be important in regulating HO following pheromone treatment, as they play an essential role in defining the regulatory properties of the HO promoter during log phase (30). The SBF binding sites at HO are embedded within nucleosomes, whereas the SBF sites at CLN2 are in a nucleosome-depleted region (NDR) (40). We made HO-CLN2 promoter chimeras in which some of the SBF binding sites at HO were now within an NDR; expression of this HO(urs2L:CLN2-wtNDR) chimera does not require Swi5, which is normally required for HO activation, establishing the ultimate importance of recruiting SBF to URS2 for HO activation (30). The absence of nucleosomes also altered the timing of promoter activation and bypassed the regulation that normally restricts expression only to mother cells. We made an additional chimera, HO(urs2L:CLN2-mutNDR), where the CLN2 sequence was altered so that nucleosomes were once again present over the SBF sites in URS2. This was possible because Bai et al. (41) identified binding sites within CLN2 that are required to create an NDR, and mutation of these sites in the CLN2-mutNDR version allowed nucleosome occupancy to be restored. In our promoter chimera experiments, the HO(urs2L:CLN2-mutNDR) construct with nucleosomes present over the ectopic CLN2 sequences showed regulatory properties much like the native HO promoter.
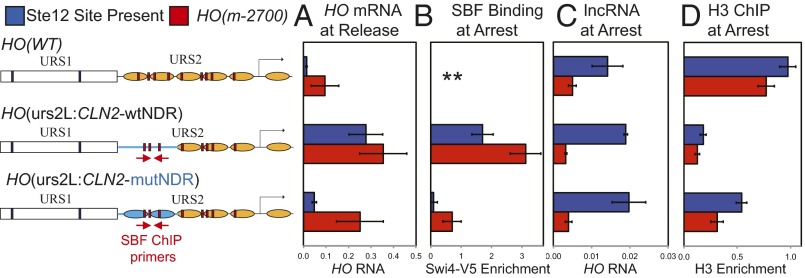
RNA was isolated from cells with the HO-CLN2 chimeras, either arrested with α-factor or at 15 min following release. Although HO mRNA is absent from wild-type cells (focusing for now on the blue bars) following release from arrest, the HO(urs2L:CLN2-wtNDR) promoter with the SBF sites within an NDR shows strong HO expression after release (Fig. 5A, line 2). In contrast, the HO(urs2L:CLN2-mutNDR) construct (line 3) in which nucleosome establishment is restored shows much weaker HO expression following release than the chimera with the NDR (line 2), but greater than HO(WT) (line 1). ChIP experiments (Fig. 5B) show strong SBF binding during arrest at the HO(urs2L:CLN2-wtNDR) promoter, whereas SBF binding at HO(urs2L:CLN2-mutNDR) was very weak, similar to that seen in wild type (Fig. 2). Importantly, the pHO-lncRNA is still expressed during the arrest in the HO(urs2L:CLN2-wtNDR) strain (Fig. 5C). An H3 ChIP experiment demonstrates reduced nucleosome occupancy in the HO(urs2L:CLN2-wtNDR) promoter, as seen previously (30). This same experiment was conducted with equivalent strains that have the HO(m-2700) mutations in the Ste12 binding site (Fig. 5, red bars). These HO(m-2700) strains with the CLN2 insertions do not express the pHO-lncRNA and show higher levels of HO expression following release, consistent with higher expression in log phase (30). These results show that an ectopic NDR can override the effects of pHO-lncRNA transcription. Thus, nucleosomes play a decisive role in determining whether SBF remains displaced from HO URS2 during α-factor arrest. Moreover, there is a correlation between the amount of SBF binding during the arrest and the ability of the promoter to express HO following release.
Fig. 5.
An NDR in the promoter allows SBF binding during arrest. The diagrams show the three promoters used, with CLN2 sequence indicated in blue, CLN2 nucleosomes represented by blue ovals, HO nucleosomes by brown ovals, Swi5 binding sites by blue boxes, and SBF sites by red boxes (see ref. 30). Six strains were used. Three of the strains had the native Ste12 site present at –2700 of the promoter (blue bars), with variations in the URS2 promoter region: HO(WT), HO(urs2L:CLN2-wtNDR), which has SBF sites within the CLN2 NDR, and HO(urs2L:CLN2-mutNDR), which has SBF sites within CLN2 sequences but has nucleosomes present. Three additional strains have nucleotide substitutions eliminating the Ste12 binding site [HO(m-2700), red bars], with the same three URS2 regions. These six strains were arrested with α-factor, and SBF binding at arrest was measured by ChIP (B), the pHO-lncRNA was measured by RT-qPCR (C), and histone H3 occupancy was measured by ChIP (D). Cells were released from arrest, and at 15 min, HO mRNA was measured by RT-qPCR (A). For the two strains with HO-CLN2 chimeric promoters, part of the CLN2 promoter was deleted and replaced with CLN1 sequence, so that the SBF ChIP primers used (indicated on the diagram) only amplify sequence from the HO-CLN2 chimeras (30). ** For HO(WT), the ChIP primers only amplified the native CLN2 promoter. In D, HO-specific primers (–927 to –828) were used for HO(WT), and CLN2-specific primers used for HO(urs2L:CLN2-wtNDR) and HO(urs2L:CLN2-mutNDR).
Discussion
One hallmark of differentiated cells is the ability to remember decisions regarding which genes should be expressed after transiting a cell cycle. Transcriptional memory at promoters can be maintained by a number of mechanisms, including DNA methylation (42), histone modification (43, 44), localization to the nuclear periphery (45, 46), and heterochromatin formation (47, 48). Both small RNAs (49, 50) and lncRNAs (14, 15) can be required for these epigenetic effects. Promoter binding by the SBF factor is part of memory at the HO promoter (8). Here we show that SBF binding and HO promoter memory can be disrupted by transcription of a lncRNA and that SBF binding sites within nucleosomes are required for this transcription-dependent loss of memory.
Regulation at HO is complex, as is the transcriptional memory. The Swi5 DNA-binding protein enters the nucleus in M phase and initiates chromatin changes at the promoter (3, 6). However, Swi5 is quickly degraded and is not present at the promoter at the time HO is transcribed in late G1 (51). The time between Swi5 binding and HO expression is normally only 20 min, but this interval can be extended substantially by nutrient starvation or by CDK inactivation. HO is still expressed when these cells reenter the cell cycle but only if Swi5 was present during the previous M phase (6). This ability of cells to remember the effects of Swi5 on the promoter has been described as memory (6), and we showed that this memory consists of SBF and Mediator bound to URS2 of the HO promoter (8).
In contrast to nutrient deprivation or CDK inactivation, HO is not expressed when cells reenter the cell cycle following G1 arrest by α-factor (11). α-Factor exposure leads to expression of a lncRNA from the HO upstream region (33, 34), and we show here that this pHO-lncRNA originates downstream of an Ste12 binding site at about –2700 bp upstream of the HO TSS. Pheromone exposure causes activation of a MAP kinase cascade resulting in phosphorylation of the Ste12 transcription factor, which then strongly binds DNA (9, 10). Our ChIP experiments show that transcription of pHO-lncRNA causes displacement of the SBF bound at HO; thus, the promoter-bound SBF appears to constitute promoter memory, and loss of SBF binding results in loss of memory. Mutating the Ste12 binding site in the HO promoter results in loss of the α-factor–induced pHO-lncRNA, retention of SBF binding despite α-factor exposure, and HO expression following release from α-factor arrest. These results suggest that pHO-lncRNA is induced by Ste12 after α-factor exposure, that transcription by pol II leads to displacement of SBF from the HO promoter, and that promoter-bound SBF is required for HO expression after release from arrest.
The SBF binding sites at HO are embedded within nucleosomes (4). Modifying the HO promoter so that the SBF sites at the left end of URS2 are in an NDR alters regulation of the promoter (30). This HO(urs2L:CLN2-wtNDR) promoter with an NDR is expressed earlier in the cell cycle, is inappropriately expressed in daughter cells, and activation is independent of the normally required Swi5 transcription factor (30). Unlike the WT HO promoter, SBF binding is seen at the mutant promoter with a stable NDR at URS2 during α-factor arrest, and HO is expressed following release. This result suggests that pol II displaces SBF but that SBF can rebind if nucleosomes are not quickly deposited after transcription. We propose that the transcribing pol II displaces SBF from the wild-type promoter, but nucleosome redisposition is rapid and efficient, preventing SBF rebinding.
Long noncoding RNAs have also been implicated in regulation of promoter activity at other genes in yeast. For example, expression of the IME1 gene that regulates entry into meiosis is blocked by expression of the upstream IRT1 lncRNA (23). This repression of IME1 by IRT1 transcription is dependent on Set2 methyltransferase and Set3 deacetylase, suggesting that histone modifications are critical here. The FLO11 gene encodes a cell wall protein regulating adhesion between cells, and its expression is controlled by a pair of lncRNAs from the upstream region (24). lncRNAs have been proposed to regulate gene induction at the PHO5 (22) and GAL loci (21, 52). A proposed mechanism involving transcriptional displacement of the Cyc8 repressor (20) is mechanistically similar to our model at HO. SER3 encodes an enzyme involved in serine biosynthesis, and exogenous serine leads to expression of the upstream SRG1 lncRNA, which prevents expression of SER3 (18). Regulation of SER3 is most similar to HO, as rapid and efficient nucleosome assembly at the SER3 promoter after transcription through the region is an essential component of normal repression. For this reason, mutations affecting histone chaperones involved in nucleosome redeposition like FACT and Spt6 reduce repression of SER3 (19). FACT and SPT6 are both required for activation of HO transcription, so we were unable to determine whether these chromatin factors are also involved in the repression following release from α-factor arrest. The DNA-binding activator for SER3 is not known, so it has not been possible to ask whether RNA polymerase displaces the SER3 activator. An experiment with a hybrid SER3 promoter containing Gal4 binding sites showed increased Gal4 binding when mutations prevent expression of the SRG1 lncRNA (19). HO may not be unique in using lncRNA transcription to modify the state of memory at a promoter.
There are good teleological reasons why a cell would not want to express HO following pheromone exposure, either because the cell is now a diploid or because it may be more successful in mating in its next cell cycle. Here we show that the cell achieves this control by using lncRNA transcription to displace the SBF memory factor from the HO promoter. Further studies will determine whether in other systems promoter-bound factors can constitute memory of prior decisions and whether factor displacement by transcription can remove this memory.
Acknowledgments
We thank Tim Formosa, Emily Parnell, and Dean Tantin for comments on the manuscript and members of the D.J.S. laboratory for helpful advice throughout the course of this project. This work was supported by National Institutes of Health Grants GM114514 and GM039067 (to N.C.E. and D.J.S., respectively). R.M.Y. was supported by NIH training Grant T32 DK007115. E.B.C. is a Howard Hughes Medical Institute postdoctoral fellow of the Jane Coffin Childs Fund. N.C.E. is a Pew Scholar in the Biomedical Sciences and Mario R. Capecchi Endowed Chair in Genetics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601793113/-/DCSupplemental.
References
- 1.Strathern JN, et al. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 2.Stillman DJ. Dancing the cell cycle two-step: Regulation of yeast G1-cell-cycle genes by chromatin structure. Trends Biochem Sci. 2013;38(9):467–475. doi: 10.1016/j.tibs.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34(4):405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarrington RM, Rudd JS, Stillman DJ. Spatiotemporal cascade of transcription factor binding required for promoter activation. Mol Cell Biol. 2015;35(4):688–698. doi: 10.1128/MCB.01285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasmyth KA. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 6.Tebb G, Moll T, Dowzer C, Nasmyth K. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 1993;7(3):517–528. doi: 10.1101/gad.7.3.517. [DOI] [PubMed] [Google Scholar]
- 7.Nasmyth K. A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985;42(1):225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, et al. Stochastic expression and epigenetic memory at the yeast HO promoter. Proc Natl Acad Sci USA. 2013;110(34):14012–14017. doi: 10.1073/pnas.1306113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pi H, Chien CT, Fields S. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol Cell Biol. 1997;17(11):6410–6418. doi: 10.1128/mcb.17.11.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 11.Nasmyth K, Seddon A, Ammerer G. Cell cycle regulation of SW15 is required for mother-cell-specific HO transcription in yeast. Cell. 1987;49(4):549–558. doi: 10.1016/0092-8674(87)90457-0. [DOI] [PubMed] [Google Scholar]
- 12.Klar AJ, Strathern JN, Hicks JB. A position-effect control for gene transposition: State of expression of yeast mating-type genes affects their ability to switch. Cell. 1981;25(2):517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- 13.Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1773(8):1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319(5859):94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet. 2013;14(12):880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 18.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429(6991):571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 19.Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011;25(1):29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cloutier SC, et al. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell. 2016;61(3):393–404. doi: 10.1016/j.molcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 2013;11(11):e1001715. doi: 10.1371/journal.pbio.1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA. 2007;104(19):8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Werven FJ, et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150(6):1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bumgarner SL, et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45(4):470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 26.Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19(8):773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- 27.Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 29.Voth WP, et al. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 2007;26(20):4324–4334. doi: 10.1038/sj.emboj.7601859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarrington RM, Goodrum JM, Stillman DJ. Nucleosomes are essential for proper regulation of a multigated promoter in Saccharomyces cerevisiae. Genetics. 2016;202(2):551–563. doi: 10.1534/genetics.115.183715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tellmann G. The E-Method: A highly accurate technique for gene-expression analysis. Nat Methods. 2006;3:i–ii. [Google Scholar]
- 32.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasmyth K. At least 1400 base pairs of 5′-flanking DNA is required for the correct expression of the HO gene in yeast. Cell. 1985;42(1):213–223. doi: 10.1016/s0092-8674(85)80117-3. [DOI] [PubMed] [Google Scholar]
- 34.Law GL, Bickel KS, MacKay VL, Morris DR. The undertranslated transcriptome reveals widespread translational silencing by alternative 5′ transcript leaders. Genome Biol. 2005;6(13):R111. doi: 10.1186/gb-2005-6-13-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: Cis- and trans-acting regulators. Cell. 1987;48(3):389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 36.Sorrells TR, Booth LN, Tuch BB, Johnson AD. Intersecting transcription networks constrain gene regulatory evolution. Nature. 2015;523(7560):361–365. doi: 10.1038/nature14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 38.Miller AM, MacKay VL, Nasmyth KA. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- 39.Mathias JR, Hanlon SE, O’Flanagan RA, Sengupta AM, Vershon AK. Repression of the yeast HO gene by the MATalpha2 and MATa1 homeodomain proteins. Nucleic Acids Res. 2004;32(22):6469–6478. doi: 10.1093/nar/gkh985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18(4):544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai L, Ondracka A, Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell. 2011;42(4):465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 43.Audergon PN, et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015;348(6230):132–135. doi: 10.1126/science.1260638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348(6230):1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21(1):127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taddei A. Active genes at the nuclear pore complex. Curr Opin Cell Biol. 2007;19(3):305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol. 2010;26:471–501. doi: 10.1146/annurev.cellbio.042308.113225. [DOI] [PubMed] [Google Scholar]
- 48.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 49.Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14(2):100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97(3):299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 52.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45(3):279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]