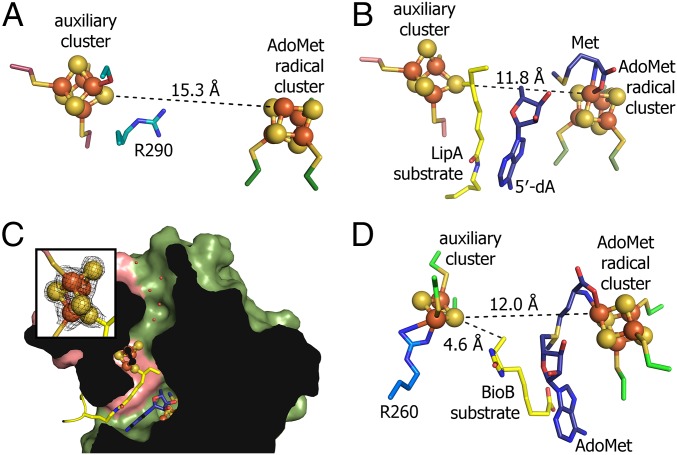
Fig. 2.
The active sites of LipA and BioB. (A) Substrate-free LipA with R290 blocking access to auxiliary cluster. (B) Intermediate-bound LipA structure. (C) A cross-section of the intermediate-bound LipA structure showing water-filled cavity created by residues in β1 (E71–T73), β2 (Y117–T119), and the C-terminal helix (S292–R297). Inset depicts simulated annealing omit density (2Fo−Fc) around the auxiliary cluster contoured at 2.0σ. (D) BioB active site (Protein Data Bank ID code 1R30) with a highly conserved arginine residue, R260, ligating the auxiliary [2Fe-2S] cluster. AdoMet ligates the AdoMet radical cluster, whereas dethiobiotin, the substrate, is between AdoMet and the auxiliary cluster.