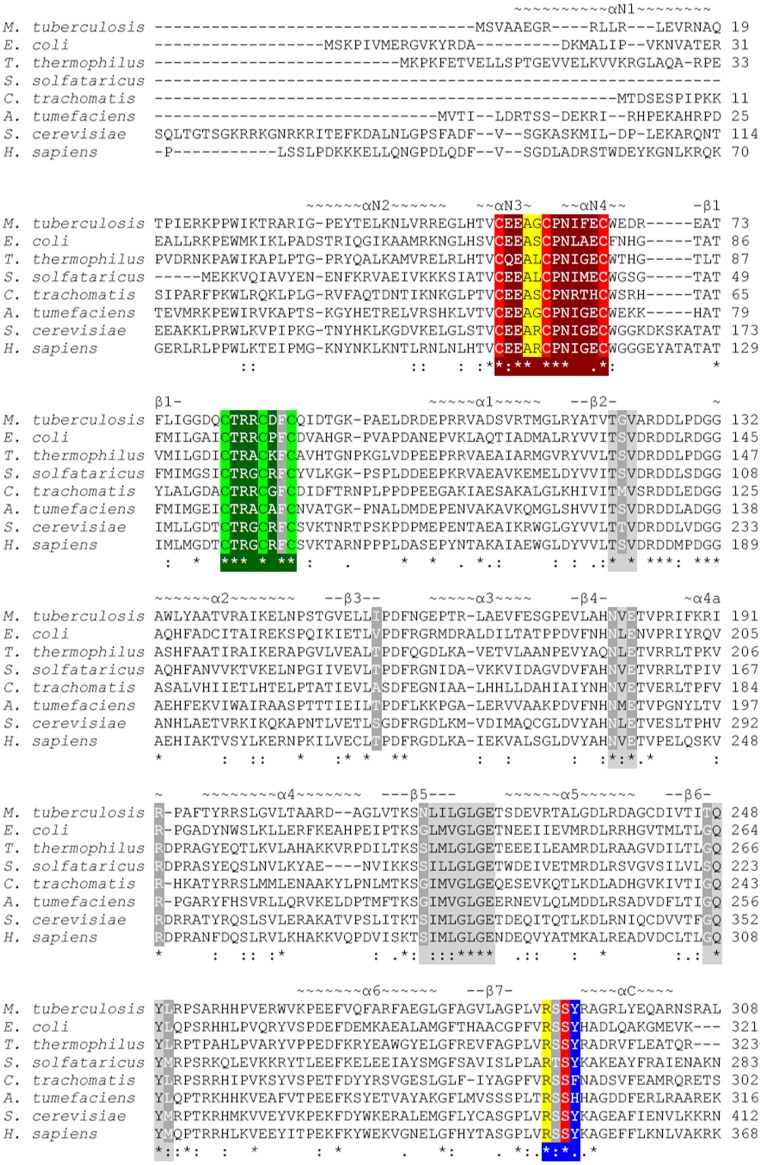
Fig. S2.
Sequence alignment of annotated LipAs from M. tuberculosis, E. coli, Thermus thermophilus, Sulfolobus solfataricus, Chlamydia trachomatis, Agrobacterium tumefaciens, Saccharomyces cerevisiae, and Homo sapiens truncated at residue 308 (M. tuberculosis numbering). Important residues are highlighted as follows: blue, R(T/S)SФ motif; dark gray, residues contacting 5′-dA and methionine; dark green, AdoMet radical CX3CXФC motif; dark red, CX4CX5C motif; light gray, canonical AdoMet binding motifs; light green, AdoMet radical cluster ligands; light red, auxiliary cluster ligands; and yellow, residues contacting the octanoyllysine residue of substrate. Secondary structure is indicated above the sequence, with N standing for N-terminal extension and C standing for C-terminal extension. A corresponding topology diagram is shown in Fig. S1.