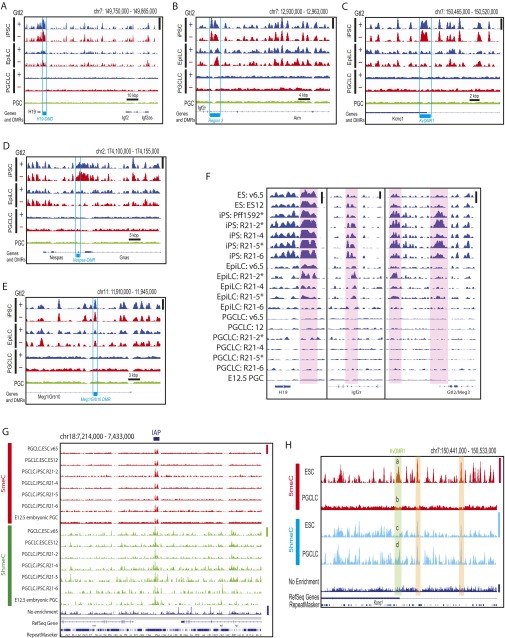
Fig. S8.
Demethylation at the ICRs in PGCLCs. Deep-sequencing tracks of 5meCs at the ICRs in the genomes of Gtl2(+) and Gtl2(−) mouse iPS cells and their EpiLC and PGCLC derivatives are shown. (A) Igf2-H19, (B) Igf2r, (C) Kcnq1, (D) Gnas, and (E) Meg1/Grb10. (F) 5meC deep-sequencing tracks of the individual cell clones at the H19, Igf2r, and Gtl2/Meg3 ICRs (shaded in red). Asterisks indicate the Gtl2(×) iPSC clones, which show abnormal hypermethylation of the Gtl2/Meg3 ICRs, and their derivatives. Height of peaks in A–F reflects relative strength of DNA methylation across all of the tracks in each panel (or each subpanel of F), linearly scaled between the baseline and the maximal methylation in the displayed area shown with vertical bars at right. (G) Retention of DNA methylation and hydroxymethylation at an IAP location in PGCLCs and PGCs. Height of peaks reflects relative strength of DNA methylation across the seven 5meC tracks (linearly scaled between the baseline and the maximal methylation in the displayed area indicated with red vertical bar at right), seven DNA hydroxymethylation (5hmeC tracks, green vertical bar), or nonenriched genome resequencing coverage (Nonenriched track, blue vertical bar). IAP location is shown by horizontal bar at the top. (H) DNA methylation and hydroxymethylation profiles of the v6.5 mouse ESCs and their PGCLC derivatives at the ICR of the Kcnq1 imprinting cluster. Height of peaks reflects relative strength of DNA methylation across the two 5meC tracks (linearly scaled between the baseline and the maximal methylation in the displayed area indicated with red vertical bar at right), two 5hmeC tracks (cyan bar), or nonenriched genome resequencing track (blue bar). The ICR (KvDMR1) is shown with green shading, in which strong DNA methylation in the ESCs (a) was lost in PGCLCs (b) whereas DNA hydroxymethylation was enhanced upon ESC differentiation to PGCLC (compare peaks c and d). Two regions shaded with orange show loss of DNA methylation upon ESC differentiation to PGCLC without significant changes in DNA hydroxymethylation.