Significance
In healthy cells, phosphatidylserine (PtdSer) is exclusively localized at inner leaflets of plasma membranes. When cells undergo apoptosis, caspases cleave a membrane protein, XK-related protein (Xkr) 8, at the C terminus, and the cleaved Xkr8 supports scrambling of phospholipids between inner and outer leaflets of plasma membranes. The PtdSer, thus exposed to the cell surface, is recognized by macrophages as an “eat me” signal for engulfment of dead cells. We report that Xkr8 is present as a complex with basigin or neuroplastin that work as a chaperone to localize Xkr8 at plasma membranes. When Xkr8 is cleaved by caspase, the complex undergoes a higher-order structure. These results will contribute to our understanding of how the Xkr8 scrambles phospholipids when cells undergo apoptosis.
Keywords: phospholipid scramblase, Xkr8, chaperone, basigin, neuroplastin
Abstract
Xk-related protein (Xkr) 8, a protein carrying 10 transmembrane regions, is essential for scrambling phospholipids during apoptosis. Here, we found Xkr8 as a complex with basigin (BSG) or neuroplastin (NPTN), type I membrane proteins in the Ig superfamily. In BSG−/−NPTN−/− cells, Xkr8 localized intracellularly, and the apoptosis stimuli failed to expose phosphatidylserine, indicating that BSG and NPTN chaperone Xkr8 to the plasma membrane to execute its scrambling activity. Mutational analyses of BSG showed that the atypical glutamic acid in the transmembrane region is required for BSG’s association with Xkr8. In cells exposed to apoptotic signals, Xkr8 was cleaved at the C terminus and the Xkr8/BSG complex formed a higher-order complex, likely to be a heterotetramer consisting of two molecules of Xkr8 and two molecules of BSG or NPTN, suggesting that this cleavage causes the formation of a larger complex of Xkr8-BSG/NPTN for phospholipid scrambling.
Phospholipids are asymmetrically distributed in plasma membranes by flippases that actively translocate phosphatidylserine (PtdSer) and phosphatidylethanolamine from the outer to inner leaflets of the membrane (1, 2). This asymmetrical distribution is disrupted in the activated platelets and apoptotic cells (3), in which the PtdSer exposed on the cell surface serves as a scaffold for blood clotting factors and as an “eat me” signal, respectively (4, 5). ATP11A and ATP11C, members of the P4-type ATPase family, act as flippases at the plasma membrane in most cells (6, 7). Two processes, flippase inactivation and scramblase activation, must occur to disrupt the asymmetrical phospholipid distribution and expose PtdSer on the cell surface (8).
Scramblases are membrane proteins that nonspecifically and bidirectionally transport phospholipids between the two plasma membrane leaflets (9). Ca2+-activated phospholipid scrambling is mediated by membrane proteins that belong to the transmembrane protein (TMEM)16 (also called ANO) family (8). Of 10 human TMEM16-family members, 5 are Ca2+-activated phospholipid scramblases at plasma membranes. TMEM16F exposes PtdSer in activated platelets and osteoblasts (10–12). The tertiary structure of fungal TMEM16 and the biochemical characterization of mouse TMEM16 family members indicate that TMEM16 forms a homodimer that directly binds Ca2+ (13).
Phospholipid scrambling and PtdSer exposure in apoptotic cells is mediated by another family of membrane proteins, the XK-related (Xkr) proteins (8). Of 10 human Xkr family members, Xkr8 (ubiquitously expressed) and Xkr4 and Xkr9 (expressed in specific tissues) are cleaved by caspase during apoptosis to expose PtdSer (14, 15), but how the cleavage activates these Xkrs to scramble phospholipids is unknown. XK, the founding member of the Xkr family, associates with Kell, a type II membrane protein (16). Whether Xkr8 and other Xkr-family members associate with other proteins has not been addressed.
In this report, we found that Xkr8 solubilized in different detergents behaved differently in blue native PAGE (BN-PAGE). We purified the Xkr8 complex from membrane fractions and determined that it associated with basigin (BSG) or neuroplastin (NPTN) (17, 18). We found that BSG or NPTN is required for Xkr8’s function as a caspase-dependent phospholipid scramblase. In apoptotic cells, the caspase-cleaved Xkr8, together with BSG or NPTN, formed a higher-order complex, suggesting that BSG and NPTN might also be involved in scrambling phospholipids.
Results
Identification of BSG and NPTN in the Xkr8 Complex.
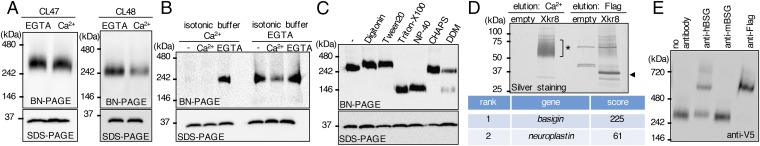
To assess molecular characteristics of the Xkr8 protein, PLB985 cells (PLB) not expressing Xkr8 (14) were transformed with Flag-tagged human Xkr8 (hXkr8). Because the stability and subunit structure of membrane proteins is often regulated by Ca2+ and detergent (19, 20), PLB-hXkr8 was lysed in different detergents (CL47 or CL48) with mild and intermediate stringency (21) containing 0.5 mM EGTA or 1.0 mM Ca2+ and separated by BN-PAGE. Western blot with anti-Flag showed that hXkr8 lysed in CL47 behaved as a large complex in the presence or absence of Ca2+ (Fig. 1A); however, when lysed with CL48 containing Ca2+, the intensity of the large hXkr8 complex decreased significantly. Because SDS/PAGE showed similar amounts of hXkr8 in CL47 or CL48 with or without Ca2+, these results suggested that native hXkr8 exists as a large complex in the absence of Ca2+, but the complex is disrupted by Ca2+ in CL48. To confirm the effect of Ca2+ on the hXkr8 complex, crude membrane fractions (Fig. S1A) were prepared from PLB-hXkr8 in isotonic buffer containing 1.0 mM Ca2+ or 0.5 mM EGTA and solubilized with CL48 containing 1.0 mM Ca2+ or 0.5 mM EGTA. As shown in Fig. 1B, when hXkr8 was prepared with the isotonic and solubilization buffers containing EGTA, it behaved as a large complex. However, there was no clear hXkr8 complex in lysates prepared with the buffers containing Ca2+, although similar amounts of hXkr8 were detected in SDS/PAGE. Adding 1.0 mM Ca2+ to the CL48 solubilization buffer severely decreased the intensity of the large complex, indicating that Ca2+ disrupted the structure of the hXkr8 complex. In agreement that different detergents have different effects on the hXkr8 complex, the treatment of CL47 lysates with Triton X-100 or Nonidet P-40, but not digitonin, Tween20, or CHAPS, reduced the size of the hXkr8 complex (Fig. 1C).
Fig. 1.
Identification of BSG and NPTN in the Xkr8 complex. (A and B) Crude membranes were prepared from PLB-hXkr8 in an isotonic buffer containing 0.5 mM EGTA (A) or 1 mM Ca2+ or 0.5 mM EGTA (B), solubilized in CL47 or CL48 (A) or CL48 (B) containing 1 mM CaCl2 or 0.5 mM EGTA, assessed by BN-PAGE or SDS/PAGE, and Western-blotted with anti-Flag. (C) Crude membranes prepared in an isotonic buffer containing 0.5 mM EGTA were solubilized in CL47. After adding the indicated detergents (0.5% for digitonin and 1% for all others), the lysates were incubated for 1 h on ice, assessed by BN-PAGE or SDS/PAGE, and Western-blotted with anti-Flag. (D) Light membranes from PLB or PLB-hXkr8 were solubilized with CL48 and applied to anti-Flag beads. Proteins were eluted with CL48 containing 1 mM CaCl2, and then with Flag peptide, separated by SDS/PAGE and silver-stained. The broad band at 60–70 kDa was analyzed by mass spectrometry (table). (E) Crude membranes from PLB expressing the N-terminally V5- and C-terminally Flag-tagged hXkr8 were lysed in CL47, incubated with anti-hBSG, anti-mBSG, or anti-Flag, analyzed by BN-PAGE, and Western-blotted with anti-V5.
Fig. S1.
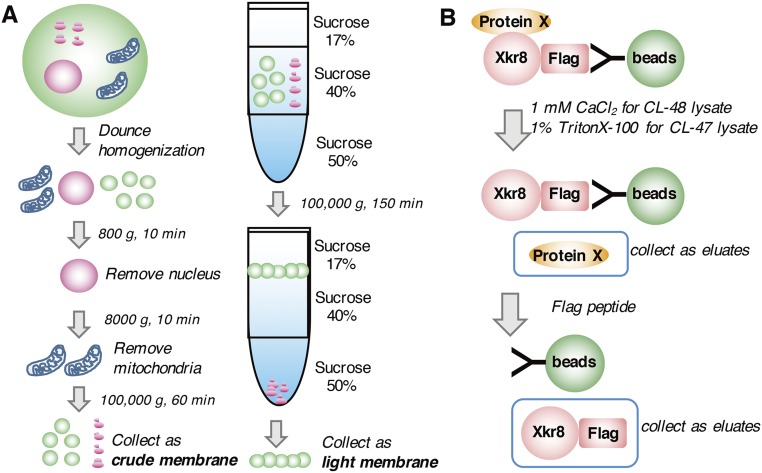
Preparation of the crude and light membrane fractions and isolation of proteins associated with hXkr8. (A) Procedure for preparing the membrane fractions. For crude membrane fractions (Left), cells were suspended in hypotonic buffer, homogenized in a Dounce homogenizer, and spun at 800 × g for 10 min at 4 °C to remove the nuclei. The supernatant was spun at 8,000 × g for 10 min to remove mitochondria, and then the crude membrane fraction was precipitated by centrifugation at 100,000 × g for 60 min. To prepare the light membrane fraction (Right), the crude membrane fraction was suspended in buffer containing 40% (wt/vol) sucrose and loaded between 17% (wt/vol) and 50% (wt/vol) sucrose layers. The light membrane fraction at the 17/40% (wt/vol) sucrose interface was collected after centrifugation at 100,000 × g for 150 min at 4 °C. (B) Procedure for isolating the proteins associated with hXkr8. The hXkr8 complex was trapped on anti-Flag beads. hXkr8- associated proteins were eluted with CL48 containing 1 mM CaCl2 or CL47 containing 1% Triton X-100. hXkr8 was released from the beads with Flag peptides.
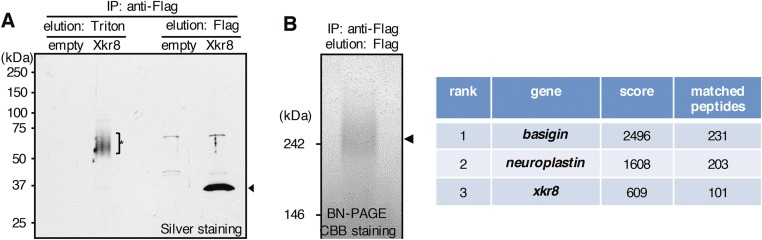
To identify molecules associated with hXkr8, light membranes were prepared (Fig. S1A) and solubilized with CL47 or CL48 containing 0.5 mM EGTA. The hXkr8 complex was trapped with anti-Flag beads, and Xkr8-bound proteins were eluted with 1 mM Ca2+ in CL48 or with 1% Triton X-100 in CL47 (Fig. S1B). In SDS/PAGE, silver staining showed broad, specific bands of 60–70 kDa in eluates with 1 mM Ca2+ (Fig. 1D) or 1% Triton X-100 (Fig. S2A). In either case, subsequent elution with Flag peptides produced a 37-kDa band that was hXkr8. Mass spectrometry indicated that the 60- to 70-kDa proteins in the Ca2+ eluates were a mixture of hBSG and hNPTN (Fig. 1D). To confirm that hXkr8 associates with BSG and NPTN in hXkr8 complex, light membranes solubilized in lauryl maltose neopentyl glycol (LMNG) were trapped with anti-Flag beads and directly eluted with Flag peptides. Staining the eluates with Coomassie Brilliant Blue (CBB) revealed a large complex on BN-PAGE (Fig. S2B), and mass spectrometry showed that hBSG, hNPTN, and hXkr8 were present in this complex. Preincubating the CL47 lysates from PLB-hXkr8 cells with anti-hBSG, but not with anti-mouse (m)BSG, shifted about half of the large hXkr8 complex to a larger band (Fig. 1E). Preincubating with anti-Flag caused a similar, but complete, shift of the large band, suggesting that the large band contained two complexes: one with Xkr8 and BSG and another with Xkr8 and NPTN.
Fig. S2.
Isolation of proteins associated with Xkr8 and the Xkr8 complex. (A) Isolation of the Xkr8-associated proteins. The light membrane fractions from PLB or PLB-hXkr8 were solubilized with CL47 and applied to anti-Flag beads. Proteins were eluted with CL47 containing 1% Triton X-100 (asterisk), and then with Flag peptide, separated by SDS/PAGE and silver-stained. (B) Isolation of the Xkr8 complex. Light membrane fractions from PLB expressing the Flag-tagged hXkr8 were lysed in LMNG and loaded onto anti-Flag beads. The hXkr8 complex was eluted with Flag peptide, separated by BN-PAGE, and stained with CBB. The broad band at 240 kDa (arrowhead) was analyzed by mass spectrometry (table).
Requirement of BSG or NPTN for Xkr8 Complex Formation.
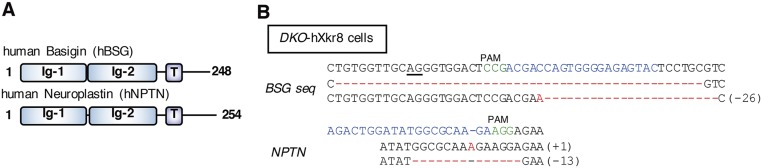
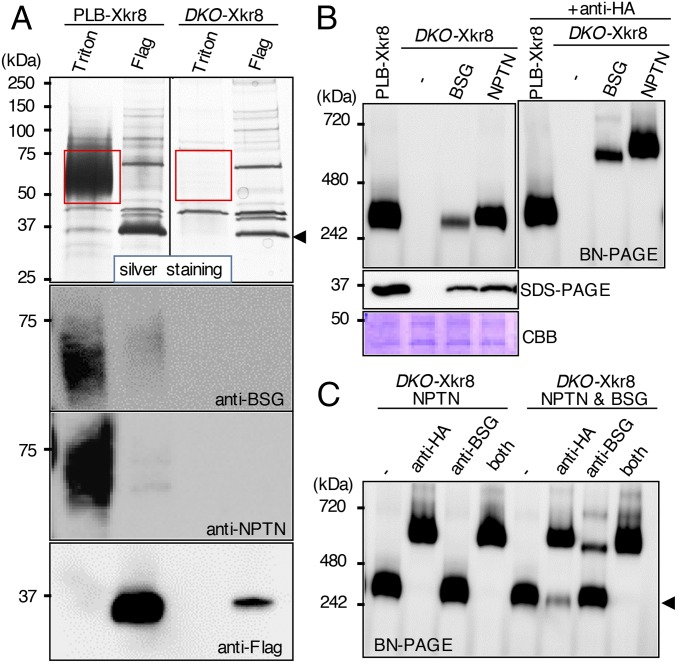
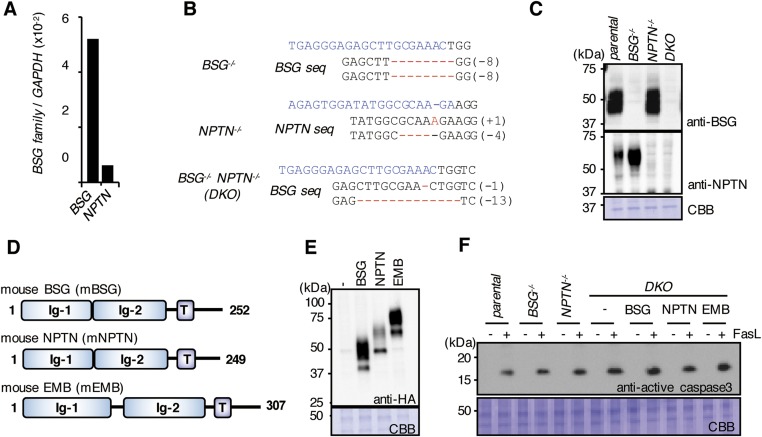
BSG and NPTN are type I membrane proteins in the Ig superfamily (17, 18) (Figs. S3A and S4). Mature hBSG and hNPTN share 39.6% identity on the amino acid sequence. We knocked out the hBSG and hNPTN genes in PLB-hXkr8 using the CRISPR-Cas system (22) (Fig. S3B). Light membrane fractions prepared from parental and BSG−/−NPTN−/−PLB (DKO)-hXkr8 were solubilized with CL47, bound to anti-Flag beads, and eluted with 1% Triton X-100 in CL47. Silver staining of parental-cell eluates showed broad bands at 60–70 kDa on SDS/PAGE that were recognized by anti-BSG or anti-NPTN (Fig. 2A). These bands were not seen in eluates from DKO-hXkr8. When subsequently eluted with Flag peptides, membrane lysates from PLB-hXkr8 released a 37-kDa protein that was recognized by anti-Flag, confirming it to be hXkr8. Unexpectedly, hXkr8 eluted from the beads was greatly reduced in membrane lysates from DKO-hXkr8, suggesting that hBSG and/or hNPTN were required to localize hXkr8 to the membrane or stabilize the Xkr8 protein.
Fig. S3.
Structure of human basigin and neuroplastin, and its editing by CRISPR-Cas system. (A) The structure of human basigin (hBSG) and neuroplastin (hNPTN) is schematically shown. Ig, Ig-like domain; T, transmembrane. (B) CRISPR-Cas–mediated editing of the hBSG and hNPTN genes. The hBSG and hNPTN target sequences (upper line) and the sequences of the mutated alleles (lower lines) in DKO-hXkr8 cells are shown. The protospacer sequence is in light blue, and the protospacer-adjacent motif (PAM) is in green. The number of added or deleted nucleotides is shown at the right. In one allele of BSG gene, 48 nucleotides including the splice acceptor site (underlined) were deleted.
Fig. S4.
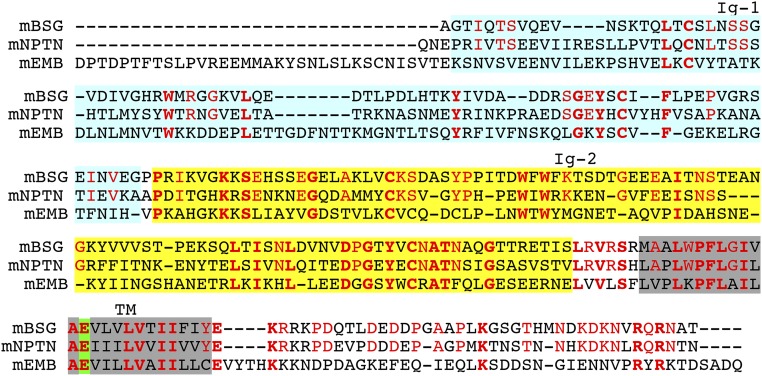
Alignment of the amino acid sequences of mouse basigin (mBSG), neuroplastin (mNPTN), and embigin (mEMB). The amino acid sequences of mBSG, mNPTN, and mEMB were aligned to obtain the maximal homology by introducing gaps (–). The amino acid residues that are identical among the three members are in bold red, and the residues that are conserved only between mBSG and mNPTN are in light red. The first (Ig-1) and second (Ig-2) Ig-like domain 1 are highlighted in light blue and yellow, respectively. The putative transmembrane regions are shadowed in gray, and the atypical glutamic acid residues in the transmembrane region are highlighted in green.
Fig. 2.
BSG or NPTN is required for the hXkr8 complex. (A) Light membranes from PLB-hXkr8 and DKO-hXkr8 were solubilized with CL47 and incubated with anti-Flag beads. Proteins were sequentially eluted with CL47 containing 1% Triton X-100 or Flag peptides, separated by SDS/PAGE, and stained with silver or Western-blotted with anti-hBSG, anti-hNPTN, or anti-Flag. Red rectangular regions indicate proteins eluted with 1% Triton X-100, and an arrowhead indicates the protein eluted by Flag peptides. (B) Crude membranes of DKO-hXkr8 transformed with HA-tagged hBSG or hNPTN were lysed with CL47, separated by BN-PAGE or SDS/PAGE, and Western-blotted with anti-Flag or stained with CBB. (Right) Membrane lysates were preincubated with anti-HA before BN-PAGE. (C) Membrane lysates from DKO-hXkr8 expressing hNPTN/HA or hNPTN/HA and nontagged hBSG were pretreated with anti-HA and/or anti-hBSG, analyzed by BN-PAGE, and Western-blotted with anti-Flag.
Accordingly, crude membrane fractions from DKO-hXkr8 did not show the large hXkr8 complex on BN-PAGE (Fig. 2B). Transforming DKO-hXkr8 with HA-tagged hBSG or hNPTN rescued the formation of large hXkr8 complex on BN-PAGE and the 37-kDa hXkr8 protein on SDS/PAGE; hNPTN was more effective than hBSG. Preincubating the CL47 membrane lysates with anti-HA shifted the complex to a larger one, confirming the presence of hBSG and hNPTN in the hXkr8 complex. To test whether the large hXkr8 complex contained hBSG, hNPTN, or both, DKO-hXkr8 cells were transformed with HA-tagged hNPTN with or without nontagged hBSG. Preincubating CL47 lysates from DKO-hXkr8-hNPTN cells with anti-HA, but not anti-hBSG, shifted the large band on BN-PAGE (Fig. 2C), confirming that the anti-hBSG did not recognize hNPTN. Preincubating the lysates from DKO-hXkr8-hNPTN-hBSG cells with anti-HA shifted 70–80% of the complex to a larger one. Preincubating the lysates with anti-hBSG shifted about 30% of the complex; preincubating with both anti-HA and anti-hBSG shifted all of complex to the larger band. These results strongly suggested that the large hXkr8 complex carried BSG or NPTN.
Effect of Mouse BSG and NPTN on Endogenous Xkr8-Mediated PtdSer Exposure.
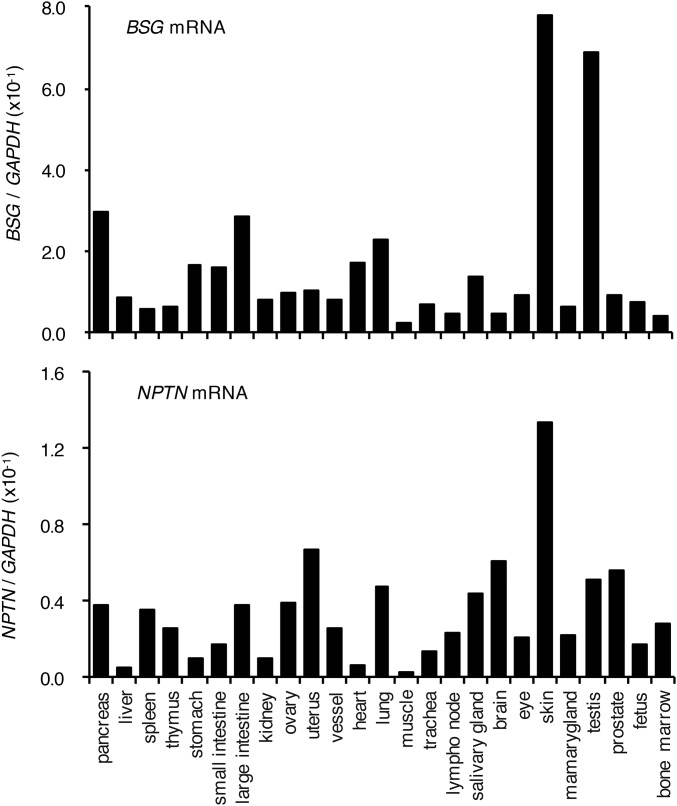
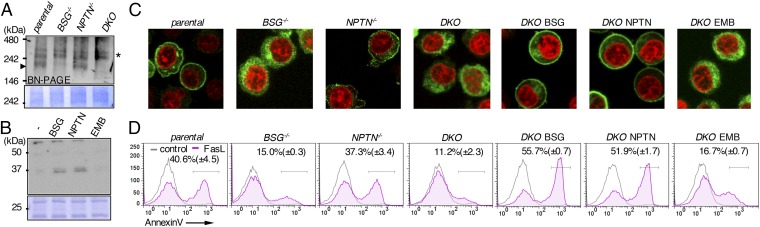
BSG and NPTN are ubiquitously expressed in various mouse tissues; in most cases the BSG mRNA level is severalfold higher than that of NPTN (Fig. S5). We next assessed the effect of mBSG and mNPTN on the endogenous Xkr8 complex. Real-time RT-PCR indicated that the mBSG mRNA level in Fas-expressing WR19L cells (WR/Fas) was about 10 times higher than that of mNPTN (Fig. S6A). We generated WR/Fas with a single or double knockout of mBSG and mNPTN genes (Fig. S6 B and C). Crude membranes of parental and mutant WR/Fas were solubilized with LMNG and separated by BN-PAGE. Western blots with anti-mXkr8 showed a large band in WR/Fas; this band was severely decreased in mBSG−/− and mBSG−/−mNPTN−/− (DKO) WR/Fas (Fig. 3B). However, the Xkr8 complex was present in mNPTN−/− WR/Fas, probably because mBSG, which was more abundant than mNPTN, contributed to the complex formation. These results suggested that mBSG or mNPTN is required for endogenous mXkr8 to form a large complex at the membrane.
Fig. S5.
Expression of basigin and neuroplastin in mouse tissues. Mouse basigin (BSG) and neuroplastin (NPTN) mRNA levels in the indicated mouse tissues were measured by real-time RT-PCR and expressed relative to GAPDH mRNA.
Fig. S6.
Expression of basigin and neuroplastin genes in WR/Fas cells, the CRISPR-Cas–mediated mutagenesis, and their effect on FasL-induced caspase activation. (A) Mouse basigin (BSG) and neuroplastin (NPTN) mRNA levels in WR/Fas cells were measured by real-time RT-PCR and expressed relative to GAPDH mRNA. (B) The mBSG and mNPTN target sequences (upper line) and the sequences of the mutated alleles of mBSG in BSG−/− and BSG−/−NPTN−/− (DKO) WR/Fas, and mNPTN in NPTN−/− WR/Fas are shown. The protospacer sequences are shown in light blue. The number of added or deleted nucleotides is shown on the right. (C) Cell lysates of crude membranes from parental, BSG−/−, NPTN−/−, or DKO WR/Fas were separated by SDS/PAGE and Western-blotted with anti-mBSG or anti-mNPTN, or stained with CBB. (D) Structures of mBSG, mNPTN, and mEMB. Ig, Ig-like domain; T, transmembrane. The number of amino acids in the mature protein is shown on the right. (E) DKO WR/Fas were transformed with HA-tagged mBSG, mNPTN, or mEMB. Cell lysates were separated by SDS/PAGE and Western-blotted with anti-HA, or stained with CBB. (F) Parental, BSG−/−, NPTN−/−, and DKO WR/Fas, and DKO WR/Fas transformed with mBSG/HA, mNPTN/HA, or mEMB/HA were treated or untreated with FasL. The cell lysates were separated by SDS/PAGE and Western-blotted with anti-active caspase 3 or stained with CBB.
Fig. 3.
BSG or NPTN is required for apoptotic PtdSer exposure. (A) Effect of BSG- and NPTN-null mutations on mXkr8 complex. Crude membranes from parental, BSG−/−, NPTN−/−, and DKO WR/Fas were solubilized with LMNG, separated by BN-PAGE, and Western-blotted with anti-mXkr8, or stained by CBB. Arrowhead, Xkr8 complex; asterisk, nonspecific. (B) Light membranes from DKO WR/Fas or its transformants expressing mBSG, mNPTN, or mEMB were lysed with LMNG, separated by SDS/PAGE, and Western-blotted with anti-mXkr8, or stained with CBB. (C) Parental, BSG−/−, NPTN−/−, and DKO WR/Fas, and DKO WR/Fas expressing mBSG, mNPTN, or mEMB were transformed with EGFP-mXkr8; cells were stained with DRAQ5 and viewed by confocal microscopy. (D) Parental, BSG−/−, NPTN−/−, and DKO WR/Fas, and DKO WR/Fas transformed with mBSG, mNPTN, or mEMB were treated or untreated with FasL, stained with Annexin V, and assessed by flow cytometry. Experiments were performed at least three times; the percentage of Annexin V-positive cells is shown with SD.
Embigin (EMB) is in the same family as BSG and NPTN (Figs. S4 and S6D). When DKO WR/Fas were transformed with HA-tagged mBSG, mNPTN, or mEMB (Fig. S6E), mBSG or mNPTN supported endogenous mXkr8’s localization to the light membranes but mEMB did not (Fig. 3B). When EGFP-tagged mXkr8 (EGFP-mXkr8) was stably expressed in WR/Fas, it was found at the plasma membrane (Fig. 3C). However, EGFP-mXkr8 localized intracellularly in mBSG−/− or DKO WR/Fas; this mislocalization in DKO WR/Fas was rescued by transformation with mBSG or mNPTN but not with mEMB. Thus, mBSG and mNPTN chaperoned mXkr8 to the plasma membrane. When parental, mBSG−/−, mNPTN−/−, and DKO WR/Fas and DKO transformants expressing mBSG, mNPTN, or mEMB were treated with Fas ligand (FasL), about 40% of WR/Fas exposed PtdSer within 1.5 h (Fig. 3D). In BSG−/− and DKO, but not NPTN−/− WR/Fas, the FasL-induced PtdSer exposure was severely decreased, although caspase 3 was activated as efficiently as in parental WR/Fas (Fig. S6F). Exogenously expressing either mBSG or mNPTN, but not mEMB, fully rescued the DKO WR/Fas’ inability to expose PtdSer, without affecting the caspase activation (Fig. 3D and Fig. S6F).
Mutational Analyses of Mouse BSG.
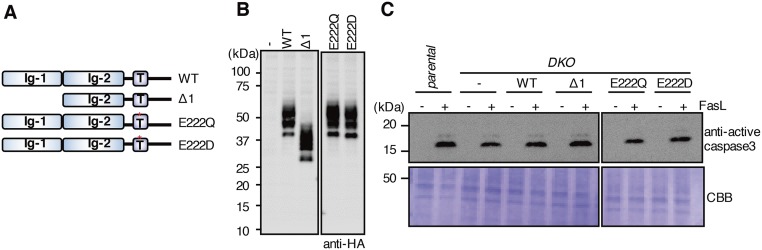
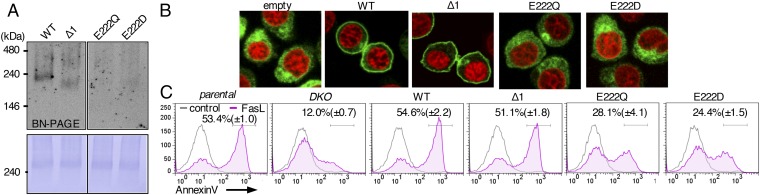
BSG and NPTN carry two Ig-like domains (Ig-1 and Ig-2) in the extracellular region. They also carry a phylogenetically well-conserved glutamic acid in the middle of the transmembrane region (at position 222 in mBSG) (Fig. S4), which is unusual for a protein with a single transmembrane region (23). To identify the mBSG region(s) involved in interacting with mXkr8, we prepared an mBSG deletion mutants (Δ1) lacking Ig-1 and the point mutants E222Q and E222D, in which the glutamic acid at position 222 was replaced by glutamine or aspartic acid (Fig. S7A). These mutants were C-terminally HA-tagged and introduced into DKO WR/Fas or DKO WR/Fas expressing EGFP/mXkr8 (Fig. S7B). A large mXkr8 complex was observed in DKO WR/Fas cells transformed by WT mBSG or Δ1 mutant, but not in cells transformed with the E222Q or E222D mutant (Fig. 4A). Similarly, transforming DKO WR/Fas-EGFP/mXkr8 with WT or Δ1 mBSG, but not the other mutants, caused EGFP/mXkr8 to translocate to the plasma membrane (Fig. 4B) and restored the DKO WR/Fas’ inability to expose PtdSer in FasL-induced apoptosis without affecting the FasL-induced caspase 3 activity (Fig. 4C and Fig. S7C). These results showed that the glutamic acid at mBSG’s transmembrane region is essential for its ability to chaperone mXkr8 to plasma membrane.
Fig. S7.
Expression of mBSG mutants in DKO WR/Fas cells and its effect on FasL-induced caspase activation. (A) Structures of WT and mutant mBSG. The mBSG Δ1 lacks the Ig-1 domain. The glutamic acid at position 222 in the transmembrane region was replaced by aspartic acid in E222D and glutamine in E222Q. Ig, Ig-like domain; T, transmembrane. Asterisk indicates aspartic acid. (B) Cell lysates from DKO WR/Fas transformants expressing WT or the indicated mutant mBSG were separated by SDS/PAGE and Western-blotted with anti-HA. (C) Effect of mBSG mutation on FasL-induced caspase activation. Parental and DKO WR/Fas and DKO transformants expressing WT or the indicated mutant mBSG were treated or untreated with FasL. The cell lysates were separated by SDS/PAGE and analyzed by Western blotting with anti-active caspase 3 or CBB staining.
Fig. 4.
Effect of mBSG mutants on apoptotic PtdSer exposure. (A) Crude membranes from DKO WR/Fas and transformants expressing WT or mutant BSG were lysed with LMNG, separated by BN-PAGE, and Western-blotted with anti-mXkr8. (B) DKO WR/Fas expressing EGFP/mXkr8 were transformed with empty vector or WT or mutant mBSG, stained by DRAQ5, and observed by confocal microscopy. (C) Parental and DKO WR/Fas and DKO transformants expressing WT or mutant BSG were treated or untreated with FasL, stained with Annexin V, and assessed by flow cytometry. Experiments were performed at least three times. The percentage of Annexin V-positive cells is shown with SD.
A Higher-Order Xkr8 Complex in Apoptotic Cells.
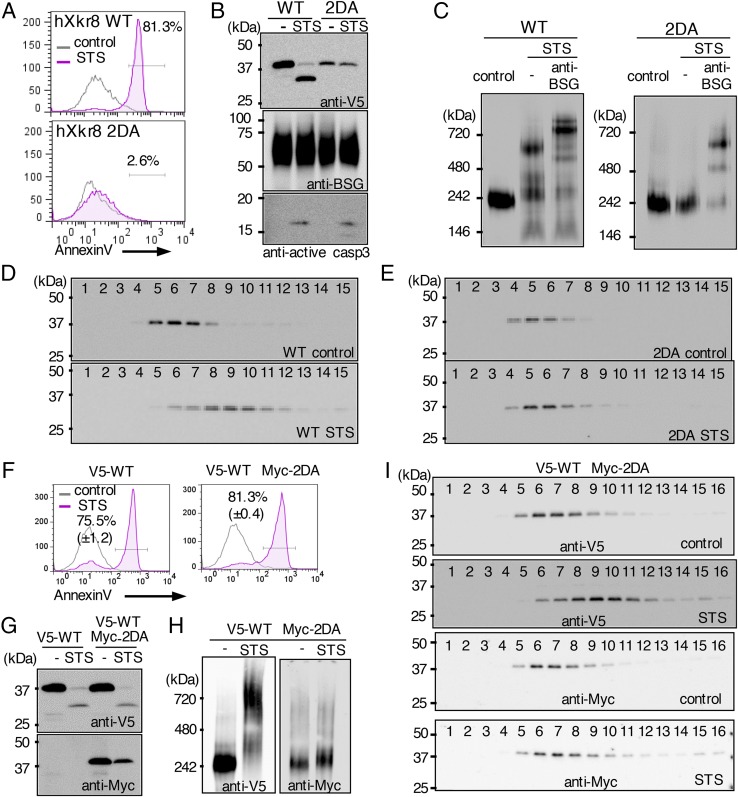
During apoptosis, Xkr8 is cleaved by caspase 3 at the C terminus to promote phospholipid scrambling (14). To assess how caspase cleavage affects the hXkr8 complex, WT and caspase-resistant (2DA) hXkr8 were N-terminally tagged with V5 and introduced into PLB. As expected, staurosporine (STS) treatment caused PtdSer exposure in cells expressing WT hXkr8, but not in those expressing the 2DA mutant (Fig. 5A). SDS/PAGE followed by Western blots with anti-V5 indicated that STS caused cleavage of WT but not 2DA hXkr8 from 37 to 30 kDa (Fig. 5B). Because BSG has no caspase-recognition sequence in the cytoplasmic region, it was not cleaved upon STS treatment. Crude membranes were lysed with LMNG and analyzed by BN-PAGE. Western blots with anti-V5 showed that a half of the large hXkr8 complex in STS-treated cells shifted to a larger band (Fig. 5C). Preincubating the membrane lysates with anti-hBSG further shifted the band to the upper region. In PLB expressing the 2DA mutant a large complex was similarly observed; this complex contained hBSG but did not shift to the larger complex when treated with STS. To confirm that the hXkr8 complex formed a larger complex in apoptosis-induced cells, membrane lysates were subjected to glycerol gradient centrifugation. CBB G-250, which reduces the aggregation of membrane proteins (24), was added to the buffer to a final concentration of 0.008%. After centrifugation, 15 fractions were collected from the top and analyzed by SDS/PAGE. Western blots with anti-V5 indicated that hXkr8 was present in fractions 4–7 from the resting PLB (Fig. 5D); hXkr8 in the STS-treated cells was recovered in the heavier fractions 7–11. However, STS did not change the sedimentation profile of the hXkr8 2DA mutant (Fig. 5E). Thus, in cells receiving apoptotic stimuli, hXkr8 was cleaved by caspase and formed a higher-order complex with BSG or NPTN.
Fig. 5.
Caspase-mediated oligomerization of the Xkr8 complex. (A–C) PLB were transformed with N-terminally V5-tagged WT or 2DA Xkr8, treated or not treated with STS, stained with Annexin V, and assessed by flow cytometry (A). Cell lysates were separated by SDS/PAGE and Western-blotted with anti-V5, anti-hBSG, or anti-active caspase 3 (B). In C, crude membranes were solubilized with LMNG, separated by BN-PAGE, and Western-blotted with anti-V5. Aliquots of lysates from the STS-treated cells were incubated 1 h with anti-hBSG on ice and applied to BN-PAGE. (D and E) The LMNG-solubilized crude membranes from STS-treated or untreated PLB expressing WT (D) or 2DA hXkr8 (E) were subjected to glycerol density gradient centrifugation. Fractions were analyzed by SDS/PAGE and Western-blotted with anti-V5. (F–I) PLB transformed with V5-tagged WT alone or cotransformed with V5-tagged WT and Myc-tagged 2DA Xkr8 were untreated or treated with STS, stained with Annexin V, and assessed by flow cytometry (F). Cell lysates were separated by SDS/PAGE and Western-blotted with anti-V5 or anti-Myc (G). In H, crude membranes were solubilized with LMNG, separated by BN-PAGE, and Western-blotted with anti-V5 or anti-Myc. In I, LMNG-solubilized membranes were analyzed by glycerol density gradient centrifugation. Fractions were analyzed by SDS/PAGE and Western-blotted with anti-V5 or anti-Myc.
We then transformed PLB with N-terminally V5-tagged WT hXkr8 alone or with Myc-tagged 2DA hXkr8. Both transformants exposed PtdSer equally in response to STS (Fig. 5F), suggesting that the 2DA mutant did not work as a dominant-negative form. Accordingly, the WT hXkr8, detected by anti-V5, was efficiently cleaved (Fig. 5G) and oligomerized after STS treatment (Fig. 5H); the 2DA mutant was not cleaved and retained the structure with the same size. Next, lysates of crude membranes from PLB transformants expressing both WT and 2DA Xkr8 were analyzed by glycerol gradient (Fig. 5I). As found with the transformants expressing WT or 2DA Xkr8, WT but not 2DA Xkr8 complex was heavier after STS treatment. These results indicated that the Xkr8 complex carried a single Xkr8 molecule, and that only the caspase-cleaved form of Xkr8 participated in formation of a larger complex. These results suggested that the active Xkr8 complex for the phospholipid scrambling is a tetramer consisting two caspase-cleaved Xkr8 and two BSG or NPTN.
Discussion
XK, a member of the Xkr family, forms a complex with Kell, a type II membrane protein with metalloproteinase activity (25). XK connects to Kell via an S–S bond at the membrane-proximal regions (16). The cysteine residue is not conserved in Xkr8 (Fig. S8), and Xkr8 does not seem to associate with Kell in mouse or human cell lines. The present study showed that Xkr8 is complexed with BSG or NPTN. Two splice forms (long and short) of BSG and NPTN (17, 18) supported Xkr8’s scramblase activity with similar efficiency. The short forms of BSG and NPTN are ubiquitously expressed. Their long forms are expressed only in photoreceptors in the retina (26) and in neurons (27), respectively, suggesting that Xkr8 is complexed with the short form of BSG or NPTN in most cells. EMB is a member of the BSG family (18), but unlike BSG and NPTN it could not chaperone Xkr8. The transmembrane region is highly conserved among BSG, NPTN, and EMB (58.3–62.5% identical) (Fig. S4). However, the second Ig-like domain is 38.4% identical between BSG and NPTN, but only 20.2% identical between BSG and EMB; this may explain why EMB does not support Xkr8’s localization to plasma membranes.
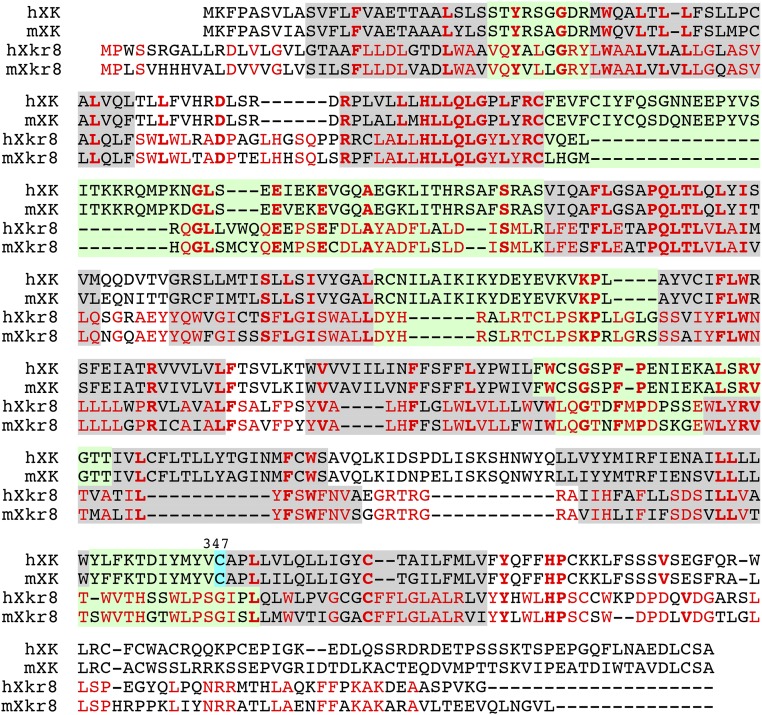
Fig. S8.
Alignment of the amino acid sequences of mouse XK and Xkr8. The amino acid sequences of human (h) and mouse (m) XK and Xkr8 were aligned to obtain the maximal homology by introducing gaps (–). The amino acid residues that are identical among the four members are in bold red, and the residues that are conserved only between human and mouse Xkr8 are in light red. The putative transmembrane regions are shadowed in gray, and the extracellular regions are in light green. The cysteine residues at position 347 of XK connected with Kell are highlighted in blue.
The apparent molecular weight of hXkr8 complexed with hBSG or hNPTN was around 240 kDa in BN-PAGE, whereas the molecular weight of the uncomplexed hXkr8 was 140 kDa. The apparent molecular weight of membrane proteins in BN-PAGE is 1.8 times higher than the actual Mr (28), suggesting molecular weight of 133 kDa for the hXkr8 complex and 78 kDa for uncomplexed hXkr8. Together with the 2DA mutant’s inability to inhibit Xkr8’s scrambling activity, these results suggest that Xkr8 (40 kDa) forms a 1:1 complex with BSG (70 kDa) or NPTN (70 kDa).
The BSG’s second Ig domain could localize Xkr8 to the plasma membrane, suggesting that BSG interacts with Xkr8’s extracellular region. Kell is a type II membrane protein with a short cytoplasmic region (29), indicating that it recognizes XK’s extracellular domain. We previously postulated that Xkr8 has six transmembrane regions (14), but reanalysis with a recently developed program (30) predicted that both XK and Xkr8 carry 10 transmembrane regions (Fig. S8). The amino acid sequences of Xkr8’s five extracellular segments have no apparent similarity with XK, which may explain why XK and Xkr8 have different partners. The extracellular loops of Xkr4 and Xkr9 that support apoptotic PtdSer (14, 15) have only limited similarity to those of Xkr8. Whether Xkr4 or Xkr9 associates with BSG or Kell family members remains to be studied.
BSG is one of the most up-regulated genes in metastatic cancer cells (31) and has multiple functions (17, 18). BSG and NPTN bind S100A9 and cyclophilins A and B and up-regulate the expression of matrix metalloproteinase (17, 32) to promote cancer-cell metastasis. BSG also laterally interacts with various proteins such as cyclophilin 60, integrins, monocarboxylate transporter 1 (MCT1), γ-secretase, and glucose transporter 1 (Glut1) (17). The glutamic acid (E222) in the BSG’s transmembrane region is dispensable for the association with MCT1 (33), suggesting that BSG interacts with various proteins via different domains. In any case, it would be interesting to examine whether basic amino acids in Xkr8’s transmembrane region interact with the glutamic acid in BSG, as found for the T-cell receptor–CD3 complex (34).
Xkr8 is cleaved by caspase to function as a phospholipid scramblase (14). Recent proteomics analyses have identified more than 1,000 caspase substrates, most of which are bystander cleavages (35). Among gain-of-function cleavages, that of kinases (ROCK1, protein kinase C, and Mst1) removes a regulatory or inhibitory domain, and the activated kinases regulate the apoptotic signaling pathway (36). Here, we found that the cleavage of Xkr8 seems to cause its conformational change for the higher-order complex formation. A similar situation is found in apoptotic DNA fragmentation in Drosophila (37); CAD (caspase-activated DNase) is complexed with its inhibitor (ICAD, inhibitor of CAD) in resting cells. When cells undergo apoptosis, both CAD and ICAD are cleaved by caspase, and CAD released from ICAD dimerizes to become an active enzyme, suggesting that the caspase cleavage of proteins in a complex can induce a conformational change that alters the complex structure. Finally, we found that Xkr8, a scramblase responsible for apoptotic PtdSer exposure, is activated by caspase-mediated formation of a large complex, likely a heterotetramer consisting of two cleaved Xkr8 and two BSG (or NPTN). Some TMEM16 family members, which also have 10 transmembrane regions and are dimers (13), support Ca2+-dependent scrambling (10). Although there is no similarity in amino acid sequence between Xkr8 and TMEM16F, they may have a similar tertiary structure. How cleaving the C-terminal tail causes the Xkr8 protein to dimerize, and how the Xkr8 and BSG (or NPTN) tetrameric complex scrambles phospholipids, will be challenging subjects for further investigation.
Materials and Methods
BN-PAGE.
A NativePAGE Novex Bis-Tris Gel System (Life Technologies) was used for BN-PAGE (24). Briefly, the solubilized membrane fraction (2.5–15 μg protein) was adjusted with sample buffer reagents [final concentrations, 50 mM Bis-Tris⋅HCl, pH 7.2, 50 mM NaCl, 10% (vol/vol) glycerol, and 0.001% Ponceau S]. For gel-shift assays, the lysates (3 μg protein) were incubated with the antibodies (0.6 μg) on ice for 1 h, loaded onto NativePAGE Novex 4–16% (wt/vol) Bis-Tris gels, and subjected to electrophoresis at 150 V for 35 min at 4 °C. The CBB G-250 concentration in running buffer was changed from 0.02 to 0.002% and the samples were further electrophoresed at 150 V for 120 min.
Preparation of Membrane Fractions.
Membrane fractions were prepared as described (15) (Fig. S1). In brief, 1–4 × 108 cells were collected, homogenized with a Dounce homogenizer in 6.5 mL of 10 mM Tris HCl buffer (pH 7.5) containing 1 mM 4-amidinophenylmethanesulfonyl fluoride (p-APMSF) and mixed with 6.5 mL of 10 mM Tris HCl buffer (pH 7.5) containing 0.5 M sucrose, 0.1 M KCl, 10 mM MgCl2, 1 mM EGTA, and 1 mM p-APMSF. After removing nuclei and mitochondria by sequential centrifugation at 800 × g for 10 min and at 8,000 × g for 10 min, crude membranes were collected by centrifugation at 100,000 × g for 1 h and solubilized by incubation at 4 °C for 3 h in 100–1,000 μL of CL47, CL48, or Buffer A [20 mM Bis-Tris⋅HCl, pH 6.8, 50 mM NaCl, 10% (vol/vol) glycerol, 1 mM p-APMSF, and cOmplete Mini] containing 0.2% LMNG. Insoluble materials were removed by centrifugation at 20,000 × g for 20 min, and the supernatants were recovered as membrane lysates. Protein concentrations were quantified by BCA Assay (Pierce).
Isolation and MS of the Xkr8 Complex.
Crude membranes from 8 × 108 PLB hXkr8 were suspended in 5.2 mL of Buffer B (10 mM Tris⋅HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, and 1 mM p-APMSF) containing 40% (wt/vol) sucrose and homogenized by passing through a 23-gauge needle. In an SW40 tube (Beckman), 3 mL of 50% (wt/vol) sucrose, 5.2 mL of membrane fractions, and 4.3 mL of 17% (wt/vol) sucrose in Buffer B were layered and centrifuged at 100,000 × g for 2.5 h at 4 °C. Light membranes at the boundary between 17% (wt/vol) and 40% (wt/vol) sucrose were diluted with Buffer B and collected by centrifugation at 100,000 × g for 1 h. Membranes were dissolved by rotating at 4 °C for 2.5 h in 1 mL of CL48 containing 0.5 mM EGTA (CL48/EGTA) or CL47 with 1 mM p-APMSF and centrifuged at 100,000 × g for 1 h. The supernatant (600 μg protein) was incubated at 4 °C for 2 h with anti-Flag agarose beads and washed three or four times with CL48/EGTA or CL47. Bead-bound proteins were eluted on ice for 2 h with 50 μL of CL48 containing 1 mM CaCl2 or CL47 containing 1% Triton X-100. The beads were washed with CL48 or CL47, and proteins were further eluted with 50 μL of CL48 or CL47 containing 160 ng/μL of 3xFLAG peptide.
For mass spectrometry, the eluates from CL48 lysates were mixed with a one-fourth volume of 5× SDS sample buffer [200 mM Tris⋅HCl, pH 6.8, 10% (wt/vol) SDS, 25% (vol/vol) glycerol, 5% (vol/vol) β-mercaptoethanol, and 0.05% bromophenol blue] and incubated at room temperature for 30 min and on ice for several days. Samples were separated by 10–20% (wt/vol) gradient SDS/PAGE (Bio Craft) and stained with a Silver Stain II Kit (Wako). Protein bands were excised and analyzed by mass spectrometry at Japan Bioservice. In some cases, light membranes were solubilized with 1 mL of Buffer A containing 0.2% LMNG and incubated with anti-Flag–conjugated magnetic beads. The beads were washed with Buffer A containing 0.2% LMNG, and bead-bound proteins were eluted on ice for 2 h with 40 μL of 160 ng/μL 3xFLAG peptide in Buffer A containing 0.2% LMNG and analyzed by BN-PAGE. The band was excised from the gel and analyzed at the Core Instrumentation Facility of Immunology Frontier Research Center at Osaka University.
Glycerol Density Gradient Centrifugation.
Crude membranes from 1.5 × 108 growing cells or 2 × 108 apoptotic cells were solubilized at 4 °C for 3 h in 0.2% LMNG-containing Buffer A (250 μL or 160 μL for growing or apoptotic cells, respectively) and centrifuged at 20,000 × g for 20 min at 4 °C. For a discontinuous glycerol density gradient, 500 μL of 27.5% (wt/vol), 500 μL of 25% (wt/vol), 700 μL of 22.5% (wt/vol), 700 μL of 20% (wt/vol), 700 μL of 17.5% (wt/vol), 700 μL of 15% (wt/vol), 600 μL of 12.5% (wt/vol), 600 μL of 10% (wt/vol), and 100 μL of 5% (wt/vol) glycerol in Buffer C (50 mM Bis-Tris⋅HCl, pH 6.8, 50 mM NaCl, and 0.008% CBB G-250) were poured stepwise into an SW55Ti tube. Solubilized membranes (50 μL containing 25–40 μg protein) were loaded on top of the gradient and centrifuged at 130,000 × g for 15.5 h at 4 °C. After centrifugation, fractions were collected from the top (eight drops each, about 300 μL) using a liquid-layer injector fractionator (Advantech) and analyzed by SDS/PAGE followed by Western blotting.
Additional information can be found in SI Materials and Methods.
SI Materials and Methods
Cell Lines, Recombinant Proteins, Antibodies, and Materials.
Human PLB985 cells (PLB) and mouse WR19L cells expressing mouse Fas (WR/Fas) were grown in RPMI1640 containing 10% (vol/vol) FCS and 50 μM β-mercaptoethanol. HEK293T cells were grown in DMEM containing 10% (vol/vol) FCS.
FasL was prepared as described (38). We used the following antibodies: rat anti-mBSG (clone RL-73) from eBioscience; goat anti-mBSG from Santa Cruz; rabbit anti-NPTN from Abcam (ab83063); mouse anti-hBSG (clone HIM6) and anti-HA (clone 16B12) from BioLegend; rabbit anti-activated caspase 3 mAb from Cell Signaling; HRP-anti-V5, HRP-anti-Myc, Alexa 488-goat anti-mouse IgG, and Alexa 488-goat anti-rat IgG from Thermo Fisher Scientific; and HRP-anti-mouse Igs, HRP-anti-goat Igs, and HRP-anti-rabbit Igs from Dako. Rabbit mAb against mXkr8 was provided by Chugai Pharmaceutical Co. HRP-labeled anti-Flag (clone M2), anti-Flag–conjugated agarose or magnetic beads, and 3xFLAG peptides were from Sigma-Aldrich. ComplexioLyte (CL)48 and CL47 were from Logopharm, LMNG from Anatrace, and DRAQ5 from BioStatus. Staurosporine was provided by Kyowa Hakko Kirin. A mixture of protease inhibitors (cOmplete, Mini, EDTA-free) was purchased from Roche Diagnostics.
cDNAs and Expression Plasmids.
The coding sequences for hBSG (GenBank accession no. D45131.1), hNPTN (NM_017455.3), mBSG (NM_001077184.1), mNPTN (NM_001293673.1), and mEMB (NM_010330.4) were prepared by RT-PCR from PLB985 (for hBSG and hNPTN) and WR19L (for mBSG, mNPTN, and mEMB) cells and verified by sequencing. The cDNA for monomeric EGFP (mEGFP) (39) was provided by M. Matsuda, Kyoto University, Kyoto. pCX4-bsr c-HA was constructed by inserting an HA tag into pCX4-bsr. To express proteins N-terminally tagged with mEGFP, or C-terminally tagged with Flag or HA, cDNAs were inserted into pMXs-puro n-mEGFP, pMXs-puro c-Flag, or pCX4-bsr c-HA, respectively. A V5- or Myc- tag was inserted into pMXs-puro hXkr8-Flag or hXkr8 2DA-Flag to generate V5- or Myc-tagged versions. In some cases, a V5-hXkr8-Flag or V5-hXkr8 2DA-Flag DNA fragment was inserted into the pNEF vector (14).
Transformation.
PLB and WR/Fas were transformed by infection with a pantropic retrovirus. Briefly, HEK293T cells were transfected with a pMX-puro vector carrying the respective cDNA, pGP for the gag-pol fusion protein (Takara Bio), and pCMV-VSV-γ-RSV-Rev for the envelope, provided by H. Miyoshi, RIKEN, Wako, Japan. The virus in the culture supernatant was concentrated by centrifugation at 6,000 × g for 16 h at 4 °C and used to infect PLB or WR/Fas. Transformants were selected with 1.5 μg/mL puromycin. PLB was transfected with Ahd I-cleaved pNEF V5-hXkr8-Flag or V5-hXkr8 2DA-Flag by electroporation using a NEPA21 (NEPAgene), subjected to a limiting dilution, and cultured with 2 mg/mL Geneticin (Gibco). The expression of hXkr8 was confirmed by Western blotting with anti-Flag mAb.
Gene Editing.
The BSG and NPTN genes were edited with the CRISPR-Cas system (22). Briefly, target sequences were selected using the CRISPR Design Tool (www.genome-engineering.org/crispr/?page_id=41). Oligonucleotides containing the target sequences were annealed and inserted into the pX330 vector (Addgene). The following oligonucleotides were used: 5′-CACCGTGAGGGAGAGCTTGCGAAAC-3′ and 5′-AAACGTTTCGCAAGCTCTCCCTCAC-3′ for mBSG; 5′-CACCGAGAGTGGATATGGCGCAAGA-3′ and 5′-AAACTCTTGCGCCATATCCACTCTC-3′ for mNPTN; 5′-CACCGGCCATCATTTCTTCTCGAAC-3′ and 5′-AAACACGACCAGTGGGGAGAGTACC-3′ for hBSG; and 5′-CACCGAGACTGGATATGGCGCAAGA-3′ and 5′-AAACTCTTGCGCCATATCCAGTCTC-3′ for hNPTN. Plasmids were electroporated into cells with a NEPA21; 4 d later, BSG−/− cells were sorted by flow cytometry with anti-BSG mAb and subjected to limiting dilution. BSG and NPTN null mutations were confirmed by sequencing the genomic DNA, and protein expression was examined by Western blots.
Real-Time PCR.
Total RNA was prepared from mouse tissues and WR/Fas cells using the RNeasy Mini Kit (Qiagen) and reverse-transcribed with SuperScript III (Thermo Fisher) or the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). The cDNAs for mBSG and mNPTN were amplified in a mixture containing LightCycler480 SYBR Green I Master (Roche Diagnostics), and the cDNA was quantified at the point where the LightCycler System detected the upstroke of the exponential phase of PCR accumulation. Linearized plasmid DNAs carrying mBSG or mNPTN cDNA were used as references. The following primers were used for real-time RT-PCR: mBSG, 5′-TCGGGAAACCATCTCACTGC-3′ and 5′-TCATGTGAGTTCCACTGCCC-3′; mNPTN, 5′-CCACCAACTCCATTGGCTCT-3′ and 5′-AGGAACCTCATCTGGCCTCT-3′.
Western Blotting.
For Western blots, proteins were transferred to a PVDF membrane (Millipore) and incubated with 5% (wt/vol) skim milk in TBST (25 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) for several hours. Membranes were probed in TBST containing 5% (wt/vol) skim milk with 6,000-fold-diluted HRP-conjugated mAb, or with mouse, rabbit, or goat Ab followed by incubation with 5,000- to 20,000-fold-diluted HRP-goat anti-mouse or anti-rabbit Ig, or HRP-rabbit anti-goat Ig. In some cases, antibodies were diluted in Can Get Signal Solution (Toyobo). Peroxidase activity was detected by the Western Lightning-ECL system (PerkinElmer), Immobilon Western Chemiluminescent HRP Substrate (Millipore), or ECL Select (GE Healthcare).
Apoptosis Induction and Flow Cytometry.
Apoptosis was induced with FasL in WR/Fas cells or with STS in PLB. In brief, 5 × 105 cells in 500 μL of culture medium were incubated at 37 °C with 11 units/mL FasL for 70–90 min or with 10 μM STS for 150 min, washed with PBS, and incubated on ice for 15 min with 1,000-fold-diluted Cy5-Annexin V in Annexin staining buffer (10 mM Hepes-NaOH, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2). Propidium iodide was added to a final concentration of 5 μg/mL, and the cells were analyzed by flow cytometry with a FACSCanto or FACSAria II.
Acknowledgments
We thank K. Saito of the DNA-chip Development Center for Infectious Diseases (Osaka University) for MS, M. Matsuda (Kyoto University) for cDNA encoding monomeric EGFP, and M. Fujii for secretarial assistance. This work was supported in part by grants-in-aid from Japan Society for the Promotion of Science (to J.S. and S.N.) and by Core Research for Evolutional Science and Technology (S.N.) and Precursory Research for Innovative Medical Care (J.S.) from Japan Agency for Medical Research and Development.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610403113/-/DCSupplemental.
References
- 1.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 2.Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim Biophys Acta. 2012;1821(8):1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: A matter of life and death. Annu Rev Physiol. 2003;65:701–734. doi: 10.1146/annurev.physiol.65.092101.142459. [DOI] [PubMed] [Google Scholar]
- 4.Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140(5):619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Lhermusier T, Chap H, Payrastre B. Platelet membrane phospholipid asymmetry: From the characterization of a scramblase activity to the identification of an essential protein mutated in Scott syndrome. J Thromb Haemost. 2011;9(10):1883–1891. doi: 10.1111/j.1538-7836.2011.04478.x. [DOI] [PubMed] [Google Scholar]
- 6.Segawa K, Kurata S, Nagata S. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases, and their regulation by caspase and calcium. J Biol Chem. 2016;291(2):762–772. doi: 10.1074/jbc.M115.690727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344(6188):1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 8.Nagata S, Suzuki J, Segawa K, Fujii T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016;23(6):952–961. doi: 10.1038/cdd.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevers EM, Williamson PL. Getting to the outer leaflet: Physiology of phosphatidylserine exposure at the plasma membrane. Physiol Rev. 2016;96(2):605–645. doi: 10.1152/physrev.00020.2015. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki J, et al. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288(19):13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468(7325):834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 12.Ehlen HW, et al. Inactivation of anoctamin-6/Tmem16f, a regulator of phosphatidylserine scrambling in osteoblasts, leads to decreased mineral deposition in skeletal tissues. J Bone Miner Res. 2013;28(2):246–259. doi: 10.1002/jbmr.1751. [DOI] [PubMed] [Google Scholar]
- 13.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516(7530):207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341(6144):403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki J, Imanishi E, Nagata S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J Biol Chem. 2014;289(44):30257–30267. doi: 10.1074/jbc.M114.583419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo D, Redman C, Lee S. Association of XK and Kell blood group proteins. J Biol Chem. 1998;273(22):13950–13956. doi: 10.1074/jbc.273.22.13950. [DOI] [PubMed] [Google Scholar]
- 17.Muramatsu T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem. 2016;159(5):481–490. doi: 10.1093/jb/mvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beesley PW, Herrera-Molina R, Smalla K-H, Seidenbecher C. The Neuroplastin adhesion molecules: Key regulators of neuronal plasticity and synaptic function. J Neurochem. 2014;131(3):268–283. doi: 10.1111/jnc.12816. [DOI] [PubMed] [Google Scholar]
- 19.Leitinger B, McDowall A, Stanley P, Hogg N. The regulation of integrin function by Ca(2+) Biochim Biophys Acta. 2000;1498(2–3):91–98. doi: 10.1016/s0167-4889(00)00086-0. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, et al. Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains. Protein Sci. 2014;23(6):769–789. doi: 10.1002/pro.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74(4):621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351(6325):414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- 24.Wittig I, Braun H-P, Schägger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 25.Khamlichi S, et al. Purification and partial characterization of the erythrocyte Kx protein deficient in McLeod patients. Eur J Biochem. 1995;228(3):931–934. [PubMed] [Google Scholar]
- 26.Ochrietor JD, et al. Retina-specific expression of 5A11/Basigin-2, a member of the immunoglobulin gene superfamily. Invest Ophthalmol Vis Sci. 2003;44(9):4086–4096. doi: 10.1167/iovs.02-0995. [DOI] [PubMed] [Google Scholar]
- 27.Langnaese K, Beesley PW, Gundelfinger ED. Synaptic membrane glycoproteins gp65 and gp55 are new members of the immunoglobulin superfamily. J Biol Chem. 1997;272(2):821–827. doi: 10.1074/jbc.272.2.821. [DOI] [PubMed] [Google Scholar]
- 28.Heuberger EH, Veenhoff LM, Duurkens RH, Friesen RHE, Poolman B. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J Mol Biol. 2002;317(4):591–600. doi: 10.1006/jmbi.2002.5416. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, Zambas ED, Marsh WL, Redman CM. Molecular cloning and primary structure of Kell blood group protein. Proc Natl Acad Sci USA. 1991;88(14):6353–6357. doi: 10.1073/pnas.88.14.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yachdav G, et al. PredictProtein--an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42(Web Server issue):W337-43. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein CA, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotechnol. 2002;20(4):387–392. doi: 10.1038/nbt0402-387. [DOI] [PubMed] [Google Scholar]
- 32.Yurchenko V, Constant S, Eisenmesser E, Bukrinsky M. Cyclophilin-CD147 interactions: A new target for anti-inflammatory therapeutics. Clin Exp Immunol. 2010;160(3):305–317. doi: 10.1111/j.1365-2249.2010.04115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoharan C, Wilson MC, Sessions RB, Halestrap AP. The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol Membr Biol. 2006;23(6):486–498. doi: 10.1080/09687860600841967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7(11):841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 35.Poreba M, Strózyk A, Salvesen GS, Drag M. 2013. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol 5(8):a008680 (abstr)
- 36.Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138(5):838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama H, et al. A novel activation mechanism of caspase-activated DNase from Drosophila melanogaster. J Biol Chem. 2000;275(17):12978–12986. doi: 10.1074/jbc.275.17.12978. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki J, Nagata S. Phospholipid scrambling on the plasma membrane. Methods Enzymol. 2014;544:381–393. doi: 10.1016/B978-0-12-417158-9.00015-7. [DOI] [PubMed] [Google Scholar]
- 39.Sadaie W, Harada Y, Matsuda M, Aoki K. Quantitative in vivo fluorescence cross-correlation analyses highlight the importance of competitive effects in the regulation of protein-protein interactions. Mol Cell Biol. 2014;34(17):3272–3290. doi: 10.1128/MCB.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]