Significance
Alzheimer’s disease is the most prevalent neurodegenerative disease still with no known cure. The disease is characterized by the development of extracellular plaques and intracellular neurofibrillary tangles. The senile plaques consist mainly of the peptide amyloid-β (Aβ) in aggregated form, called amyloid fibrils. It is believed that the Aβ amyloid fibrils play an important role in disease progression and cell-to-cell transmissibility, and small Aβ oligomers are often assumed to be the most neurotoxic species. Here, we determined the 3D structure of a disease-relevant Aβ(1–42) fibril polymorph combining data from solid-state NMR spectroscopy and mass-per-length measurements from EM. The 3D structure is composed of two molecules per fibril layer, forming a double-horseshoe–like cross–β-sheet entity with maximally buried hydrophobic side chains.
Keywords: amyloid, solid-state NMR, Alzheimer’s disease, protein structure
Abstract
Amyloid-β (Aβ) is present in humans as a 39- to 42-amino acid residue metabolic product of the amyloid precursor protein. Although the two predominant forms, Aβ(1–40) and Aβ(1–42), differ in only two residues, they display different biophysical, biological, and clinical behavior. Aβ(1–42) is the more neurotoxic species, aggregates much faster, and dominates in senile plaque of Alzheimer’s disease (AD) patients. Although small Aβ oligomers are believed to be the neurotoxic species, Aβ amyloid fibrils are, because of their presence in plaques, a pathological hallmark of AD and appear to play an important role in disease progression through cell-to-cell transmissibility. Here, we solved the 3D structure of a disease-relevant Aβ(1–42) fibril polymorph, combining data from solid-state NMR spectroscopy and mass-per-length measurements from EM. The 3D structure is composed of two molecules per fibril layer, with residues 15–42 forming a double-horseshoe–like cross–β-sheet entity with maximally buried hydrophobic side chains. Residues 1–14 are partially ordered and in a β-strand conformation, but do not display unambiguous distance restraints to the remainder of the core structure.
Alzheimer’s disease (AD) is the most prevalent neurodegenerative disease that still has no known cure and increasing incidence. The disease is characterized by the development of extracellular plaques and intracellular neurofibrillary tangles. The senile plaques consist mainly of amyloid beta (Aβ) fibrils (1, 2). The Aβ peptide is generated from the amyloid precursor protein (APP) by the proteolytic activities of β- and γ-secretase (3). The amyloid fibrils of Aβ are a pathological hallmark of AD disease and may play a central role in cell-to-cell transmissibility (reviewed in ref. 4), although their precise role in toxicity is unclear, rather with an oligomeric entity of Aβ to be most neurotoxic (5). The structure of Aβ fibrils is therefore important for a detailed understanding of the aggregation process and a molecular understanding of the progression of the disease, as well as for the development of therapeutic and diagnostic approaches.
The Aβ(1–42) fragment is the dominant Aβ species in the amyloid plaques of AD patients (6–9). Although a generalized discussion of the biological activity of Aβ is complicated by the presence of different polymorphs (10), Aβ(1–42) typically displays a higher propensity to form amyloid fibrils in vitro (11–13) and appears to be a more toxic species than Aβ(1–40). Most high-resolution structural studies have been performed on Aβ(1–40) amyloid fibrils (14–17). In addition to low-resolution structural information on Aβ(1–42) fibrils from methods such as mutagenesis studies, H/D exchange measurements, EM, solid-state NMR, and X-ray fiber diffraction (18–26), recently, a solid-state NMR-based structural model based on 11 long-range distance restraints became available (27). It shows a single Aβ(1–42) molecule within a protofilament of the fibril comprising a double horseshoe-like cross–β-sheet structure. Interestingly, the familial Osaka mutant of Aβ(1–40) amyloid fibril shows a similar fold (17). Here, we determined the 3D solid-state NMR structure of a disease-relevant polymorph of Aβ(1–42) amyloid fibrils at atomic resolution. A first model was built based on a collection of a total of 81 manually identified NMR distance restraints, as well as angular restraints and scanning transmission EM (STEM)-based mass-per-length (MPL) measurements. A final calculation based on 632 automatically identified NMR distance restraints resulted in a structure with a backbone rmsd of 0.89 Å for residues 15–42.
Results
Screening of Conditions to Obtain a Sample with One Dominant Polymorph.
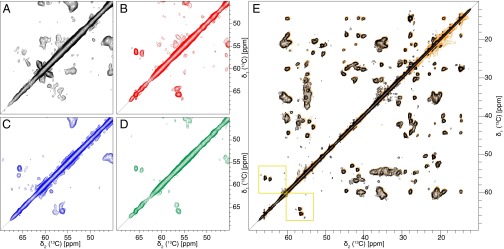
Biophysical investigations of Aβ amyloids often indicate the presence of several polymorphs within a single sample. On the mesoscopic scale, polymorphs distinguish themselves by the amount of twisting observed by EM, the number of filaments per fibril, and the diameter and MPL of the fibrils (28). Short Aβ amyloid peptides were also shown to form polymorphic microcrystals comprising different classes of steric zippers (29). Differences on the atomic scale are best detected by solid-state NMR spectra where different fibril polymorphs are distinguished by different chemical-shift fingerprints and mixtures of polymorphs by the appearance of more than one resonance line per spin probe (10, 30, 31). An NMR-guided optimization of the fibrillization conditions using seeding steps was performed to yield samples containing almost exclusively a single polymorph as indicated by the presence of a single set of resonances for all visible residues (Fig. 1 and SI Appendix, Fig. S1). In a first trial, 13C,15N-labeled Aβ(1–42) was fibrillized in phosphate buffer (condition 0, SI Appendix, Table S1). Many more cross-peaks than expected were observed in a 2D [13C,13C] dipolar-assisted rotational resonance (DARR) NMR spectrum (32, 33) as highlighted in Fig. 1A and SI Appendix, Fig. S1, which shows the serine-containing part of the 2D spectrum (indicated by a yellow square in the full aliphatic region spectrum of Fig. 1E) (an NMR acronym list is provided at the end of SI Appendix). Aβ(1–42) comprises two serines and concomitantly two 13Cα–13Cβ cross-peaks in the serine region of the spectrum are expected, but in this sample at least six cross-peaks were observed (Fig. 1A), indicating the presence of at least three polymorphs. Using 100 mM sodium chloride (NaCl) and seeding decreased the polymorphism in the sample dramatically. However, six serine peaks were still observable (Fig. 1C). In contrast, Aβ(1–42) at concentrations of either 30 or 100 μM in phosphate buffer with 100 mM NaCl and 100 μM ZnCl exhibited a single set of cross-peaks, as demonstrated for the two serine cross-peaks assigned to S8 and S26 (Fig. 1 B and D and SI Appendix, Table S1, conditions 2 and 4). Finally, condition 2 was chosen for further studies because seeds prepared under identical buffer conditions yielded reproducible solid-state NMR spectra of high quality without batch-to-batch variability (Fig. 1A). Fibrils treated with heparin (condition 1, SI Appendix, Table S1), although clearly visible by EM, did not sediment by centrifugation, and therefore no spectra could be recorded. The spectra of Fig. 1 highlight the power of NMR for the selection of conditions that lead to homogeneous samples.
Fig. 1.
Screening of conditions toward a sample with a single polymorph and reproducibility of the sample preparation. The 2D [13C,13C] DARR spectra (20-ms mixing time, 13 kHz MAS, 14 T B0) of Aβ(1–42) fibrils grown at different conditions. A–D show the serine region used to determine the amount of polymorphs in the sample. Aβ(1–42) contains two serines (residues 8 and 26), but under condition 0 (A), at least six serines can be counted. Furthermore, under this condition, the lines are very broad, and no clear defined peaks are observable. Under condition 3 (C) the lines are narrower, but still about 6 serine peaks are visible. For conditions 2 (B) and 4 (D) a single set of resonances is observed, which indicates the presence of only one morphology. (E) Superposition of two 2D [13C,13C] DARR spectra of Aβ(1–42) fibrils grown under condition 2. The spectrum in orange is from the initial sample of the screening, and the spectrum in black is the sample produced subsequently. The two spectra overlay in all regions, showing high structural similarity, and thus the sample preparation is regarded to be reproducible. In addition, the Ser region is indicated by yellow squares.
The Selected Polymorph of Aβ(1–42) Amyloids Is Disease-Relevant.
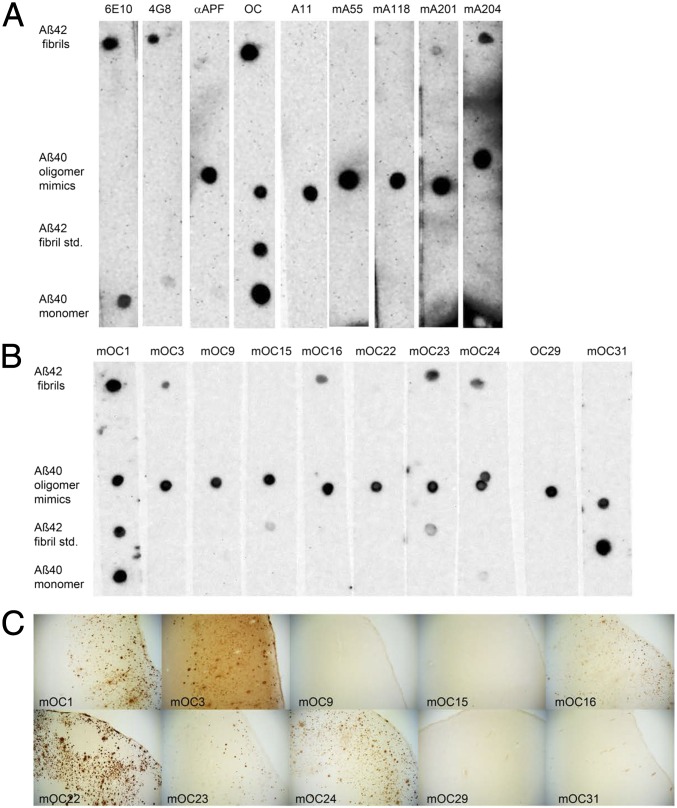
Although the selected polymorph of Aβ(1–42) fibrils is prepared at physiological pH, temperature, and salt concentration, its disease relevancy is not evident. To show this, we made use of a conformation-specific monoclonal antibody library of the C.G.G. laboratory, which is able to detect and distinguish various Aβ entities derived both from in vitro and in vivo sources (34–36). The corresponding dot blot analysis with the Aβ(1–42) fibrils prepared under condition 2 was negative for the oligomer-specific antibodies (mA55, mA118, and mA201), but positive for mA204, which indicates that the sample lacks oligomers (note that the positive staining of mA204 suggests either the presence of some special oligomers or that mA204 is not entirely oligomer-specific). Most interestingly, in contrast to the Aβ(1–42) standard fibrils used by the C.G.G. laboratory, the NMR sample was positive to all but one fibril-specific antibody (i.e., mOC1, mOC3, mOC16, mOC23, and mOC24), which are binding intracellular deposits and senile plaques in brains of human AD patients (Fig. 2), and was negative for all of the fibril-specific antibodies (i.e., mOC9, mOC15, mOC22, mOC29, and mOC31) that are not able to detect intracellular deposits and senile plaques in human Alzheimer’s patients (Fig. 2). This finding suggests that the selected polymorph of Aβ(1–42) fibrils is disease-relevant.
Fig. 2.
Immunological characterization of the Aβ(1–42) fibrils. (A) Dot blot results of the Aβ(1–42) fibrils used for the solid-state NMR experiments show that the fibrils are OC+ and A11−. In addition to the solid-state NMR-analyzed Aβ(1–42) fibrils, Aβ(1–40) prefibrillar oligomer mimics, an Aβ(1–42) fibril standard prepared in the C.G.G. laboratory, and Aβ(1–40) monomers were spotted as positive controls for antibodies on each membrane. The Aβ samples on each membrane were then probed with one of the anti-Aβ antibodies (6E10 and 4G8), antiamyloid antisera (i.e., αAPF, A11, and OC), or antioligomer monoclonal antibodies (mAs) (mA55, mA118, mA201, and mA204). The 6E10 and 4G8 are commercially available anti-Aβ antibodies. Amyloid conformation specific antisera, αAPF, A11, and OC, are specific for annular protofibrils (APF), prefibrillar oligomers (A11), and fibrils (OC), respectively. The results show that the Aβ(1–42) fibrils used for the solid-state NMR analysis (top row) were immunoreactive with 6E10, 4G8, and OC, but not αAPF, A11, or by the A11-derived monoclonal antibodies mA55 and m118. The Aβ(1–42) fibrils were positive for mA201 (weakly) and mA204. (B) The Aβ(1–42) fibrils used for the solid-state NMR analysis show reactivity to a subset of selected OC-derived fibril-specific monoclonal antibodies (mOCs). In addition to the fibril sample, the three different Aββ samples in A were probed with 10 mOC monoclonals. The Aβ(1–42) fibrils were immunoreactive with mOC1, mOC3, mOC16, mOC23, and mOC24, but not by mOC9, mOC15, mOC22, mOC29, and mOC31. They are immunologically distinct from the Aβ(1–42) fibril standard made under different conditions that reacts weakly with mOC15, mOC23, and strongly with mOC31. (C) All of the mOC antibodies that showed reactivity with the Aβ(1–42) NMR fibrils from B also stain plaques in human AD brain (mOC 1, mOC3, mOC16, mOC23, and mOC24). Most of the antibodies that fail to react with the Aβ(1–42) NMR fibrils also fail to stain plaques in human brain (mOC9, mOC15, mOC29, and mOC31). mOC22 fails to stain the Aβ(1–42) NMR sample, but stains plaques in human brain. (Magnification: 40×.)
MPL Measurements to Obtain the Number of Monomers per Layer.
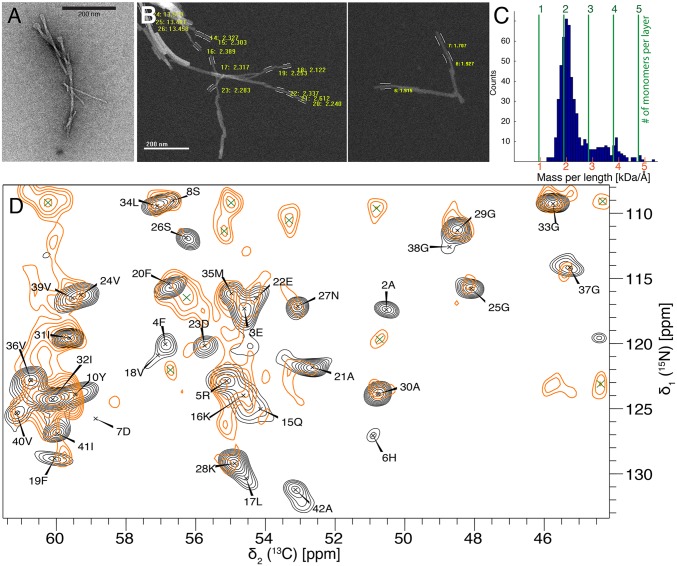
One important parameter necessary for the structure determination of an amyloid fibril is the MPL measurement using STEM (37–39). Combined with the knowledge that Aβ(1–42) amyloid fibrils are composed of an in-register intermolecular parallel cross–β-sheet entity (see below) with a repetition rate at every 0.48 nm (i.e., the distance between two β-strands across the β-sheets), the number of molecules per layer (i.e., per 0.48 nm) can be elucidated. The MPL measurements were performed on two Aβ(1–42) amyloid fibrils samples coming from two independent batches (i.e., one sample was 15N,13C-labeled and the other one unlabeled). Both samples were evaluated separately (with a MWtheor = 4,772 Da for the 15N,13C-labeled sample, and a MWtheor = 4,517 Da for the unlabeled sample) with undistinguishable results, allowing for a combination of the data. A total of 537 MPL measurements were performed manually on single filaments, which were distinguished from bundled fibrils by eye. The analyzed data show a clear peak at an MPL value of 1.9 kDa/Å, corresponding to the presence of two Aβ(1–42) molecules per 0.48 nm (Fig. 3 A–C). A small peak at MPL of 3.8 kDa/Å (corresponding to four molecules per 0.48 nm) can be attributed to a lateral stacking of two filaments. Because only a single set of cross-peaks is present in the solid-state NMR spectra (40), the two molecules must be related by a twofold symmetry. Other MPL studies on Aβ(1–40) amyloid fibrils showed either two or three molecules per unit length (15–17).
Fig. 3.
NMR and STEM measurements of the Aβ(1–42) fibrils highlighting the presence of two symmetric Aβ(1–42) molecules per fibril layer with an in-register parallel β-sheet architecture. (A and B) Negatively stained TEM (A) and two STEM images (B) of different parts of the sample, separated by white line, of unstained, freeze-dried Aβ(1–42) fibrils. Only well-defined nonoverlapping parts, often toward the ends of the fibrils, are used for the MPL measurements and are marked with white lines. (C) Result of the MPL experiment with number of measurements with a given MPL as indicated. The number of monomers per layer of a cross–β-sheet fibril is highlighted with green dotted lines. The data indicate two monomers per layer. (D) A superposition of the NCA spectrum of uniformly 13C,15N-labeled fibrils (plotted in black) with the PAIN spectrum of mixed 15N- and 13C-labeled fibrils (plotted in orange). The superposition of cross-peaks indicates an in-register parallel β-sheet structure. The assignment of the individual cross-peaks in the NCA spectrum are given with single amino acid letter codes. Green crosses indicate i ± 1 correlations.
Secondary Structure Determination of Aβ(1–42) Amyloid Fibrils.
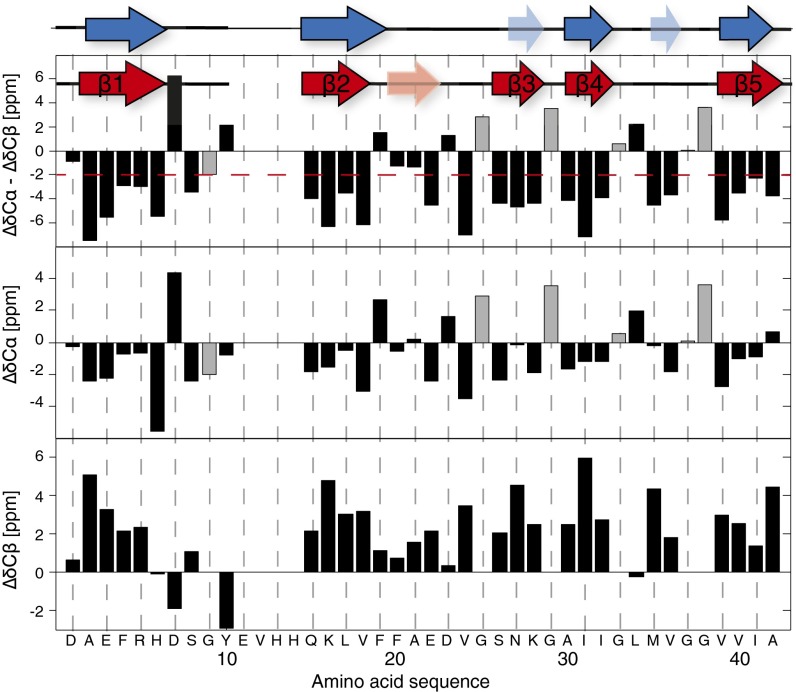
The solid-state NMR sequential backbone and side-chain resonance assignment was obtained by a suite of 2D and 3D triple-resonance experiments and is described, in detail, in an assignment note (40). The shifts have been deposited in the Biological Magnetic Resonance Bank (BMRB) database (accession no. 26692). All residues were assigned, except residues 10–14, which seem to be dynamic. The sequential assignment allows for an identification of the secondary structure of the biomolecule by the analysis of the so-called secondary chemical shifts (ΔδCα and ΔδCβ): that is, the difference between measured 13Cα (13Cβ) chemical shifts and corresponding random coil 13Cα (13Cβ) chemical shifts (38). Five β-sheets—residues 2–6 (β1), 15–18 (β2), 26–28 (β3), 30–32 (β4), and 39–42 (β5)—could be identified by using secondary chemical shifts (Fig. 4). The use of the TALOS+ databank approach (41) led to very similar sheet locations (Fig. 4, blue secondary-structure elements). Next, a [13C,15N]-proton-assisted insensitive nuclei (PAIN) experiment (42) was measured on a mixed sample of exclusively 13C-labeled and exclusively 15N-labeled Aβ(1–42) in a 1:1 ratio. This spectrum is compared with a NCA spectrum of a uniformly 13C,15N-labeled sample. At most of the intraresidual 13C-15N cross-peaks identified in the NCA spectrum and labeled with black letters in Fig. 3D, we also found peaks in the [13C,15N]-PAIN spectrum of the mixed-labeled sample, indicating the presence of in-register parallel β-sheets for β-strands β2–β5. A noticeable exception are residues 1–10, which lack cross-peaks in the mixed-labeled [13C,15N]-PAIN and also in the mixed-labeled [13C,15N]-transferred-echo double-resonance (TEDOR) spectra (SI Appendix, Fig. S2) (43, 44) and thus for residues from β-strand β1 (i.e., D1–Y10) no information about the register could be obtained. There are also no cross-peaks in the [13C,15N]-PAIN and -TEDOR spectra recorded on the mixed-labeled sample for residues L17, D23, M35, I41, and A42, most of which are not located in β-strands (Fig. 3 and SI Appendix, Fig. S2).
Fig. 4.
Secondary structural elements of the Aβ(1–42) fibrils derived from secondary chemical shifts and from TALOS+. The secondary chemical shifts (ΔδC), which is the difference between measured 13C chemical shifts and corresponding random coil 13C values, are listed vs. the amino acid sequence for 13Cα and 13Cβ, respectively. Stretches of three continuous residues with ΔδCα – ΔδCβ < –2 ppm were identified and used as an indication of β-sheet secondary structure. The resulting five β-strands are highlighted on top by red arrows. The pale red arrow denotes three additional negative (ΔδC) in a row, but not fulfilling the ΔδCα – ΔδCβ < –2 ppm criterion. Alternatively, a TALOS+ analysis was performed, and the predicted five β-strands are indicated with blue arrows. Two stretches with only two residues in the β-strand conformation are indicated in pale blue. The glycines Cα shift is shown in gray.
To strengthen further the finding that the N-terminal segment comprising residues 1–13 appears to be overall less structured, limited proteolysis with proteinase K and trypsin analyzed by solution-state NMR was performed (SI Appendix, Fig. S3). Proteinase K and trypsin are both cleaving between R5 and H6 in the N-terminal region, and thus the results for both proteases were similar, with a somewhat less prominent digestion by trypsin. As expected, the lower signal intensities of the N-terminal residues D1–S8 upon limited proteolysis compared with the corresponding signals from an untreated samples indicates that the N-terminal segment is in part structurally disordered and/or solvent-accessible. However, it is also partly protected, because a total signal loss was only achieved by a proteinase K treatment at very high proteinase K concentration [Aβ(1–42) ratio of 1:10 (wt:wt)] and overnight incubation at 37 °C (SI Appendix, Fig. S3). In summary, the N-terminal 14 residues of the Aβ(1–42) polymorph studied here are in part structured and in part dynamic.
The 3D Structure Determination of Aβ(1–42) Amyloid Fibrils.
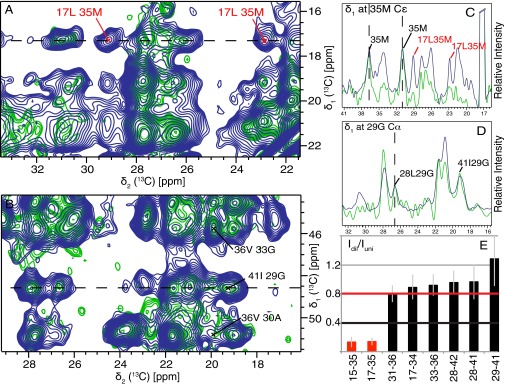
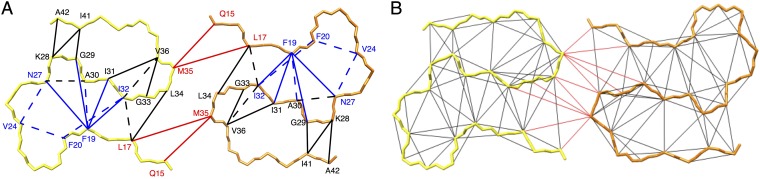
For a first 3D structural model, the following restraints were used. (i) The measure of two molecules per layer (STEM) and the presence of a single set of resonances positions two symmetrically equivalent Aβ(1–42) molecules per subunit. (ii) Dihedral angle restraints were applied, either as generic restraints in the range –200° ≤ φ ≤ –80° and 40° ≤ ψ ≤ 220° (45), for the residues involved in a β-sheet as defined by the red arrows in Fig. 4. For the presence of a β strand we request three residues in a row with a chemical-shift difference of at least −2 ppm (46, 47) (i.e., residues 2–6, 15–18, 26–28, 30–32, and 39–42) (Fig. 4). Alternatively, TALOS+ can be used to determine the position of the β-strands. As shown in Fig. 4, the results are practically identical. (iii) Intermolecular hydrogen bond restraints (2 × 32) were implemented for residues in these five β-strands. (iv) The 81 either spectrally unambiguous medium- and long-range distance restraints or restraints with low spectral ambiguity (see, for example, Figs. 5 and 6A and SI Appendix, Table S2) identified in 400 ms DARR (SI Appendix, Fig. S4), 400 μs CHHC (48) (SI Appendix, Fig. S5), and 8 ms proton assisted recoupling (PAR) (49) (SI Appendix, Fig. S6) (48). These restraints are classified as intermolecular or intramolecular (or ambiguous in this respect) according to the intensity ratio between a uniformly 13C,15N-labeled sample and a diluted sample containing uniformly 13C,15N-labeled Aβ(1–42) and nonlabeled Aβ(1–42) in a ratio of 1:3. Corresponding traces of the individual spectra of the two samples were compared by scaling nonoverlapping intraresidue or sequential peaks in the fully labeled and the diluted sample to the same intensity (Fig. 5 C and D). Three spectrally unambiguous intermolecular cross-peaks, defining contacts between two pairs of residues (Q15–M35 and L17–M35) could thus be identified, and seven intramolecular ones defining contacts between six residue pairs (Figs. 5E and 6A and SI Appendix, Figs. S4–S6 and Table S2). Representative traces showing the peak attenuation upon isotopic dilution are shown in SI Appendix, Fig. S7. A detailed analysis of all of the restraints can be found in SI Appendix, Figs. S4–S7 and Table S2. With this set of restraints, a manual 3D structure composed of 2 × 3 Aβ(1–42) molecules was calculated with CYANA (50) using upper distance restraints of 5.5, 7, and 7 Å for CHHC, PAR, and PAIN, respectively. For ambiguous peaks, SI Appendix, Table S2 indicates which of the possible restraints are fulfilled. The resulting manual fibril structure is shown in SI Appendix, Fig. S8. We note the somewhat unusual inside orientation of both F19 and F20. This feature was further verified by the presence of an additional weaker, but low-ambiguous, peak between F20 and I32 (ambiguous with I31), as shown in SI Appendix, Fig. S9.
Fig. 5.
Extracts of NMR spectra that distinguish intramolecular from intermolecular contacts. (A and B) Superposition of 2D PAR spectra recorded on a uniformly 15N,13C- labeled sample (blue contours) and on a sample of 25% uniformly 15N,13C-labeled Aβ(1–42) and 75% unlabeled Aβ(1–42) (green contours). (C and D) Selected and indicated cross-sections of the PAR spectra shown in A and B, respectively, are displayed. Corresponding spectral traces of the individual spectra of the two samples were compared by scaling the intraresidue or sequential peak intensity (for nonoverlapping peaks) for the diluted sample to the one of the uniformly labeled one. (E) Intensity ratios of cross-peaks from the diluted and uniformly labeled samples. Intermolecular and intramolecular correlations, expected to be attenuated to 25% and not attenuated, are shown as red and black bars, respectively. The full statistics are shown in SI Appendix, Fig. S5. (C) Trace extracted at the M35 Cε-resonance from the PAR spectra of uniformly (blue) and diluted (green) labeled samples. All cross-peaks of this resonance are of intermolecular nature. (D) Trace extracted at the G29 Cα-resonance from the PAR spectra of uniformly (blue) and diluted (green) labeled samples. All cross-peaks of this resonance are of intramolecular nature. Cross-peaks used to scale the two spectra are marked in C and D. Cross-peaks were classified as intramolecular contacts if the intensity ratio Idil/IUni > 0.8 with a SD margin of >0.4. Cross-peaks were classified as intermolecular contacts if the intensity ratio Idil/IUni < 0.4 with a SD margin of <0.8. No classification was done in any other cases. Details on the experimental parameters and conditions are given in SI Appendix, Fig. S5.
Fig. 6.
(A) Distance restraints used for the manual structure calculations are plotted onto the final 3D structure. Distance restraints between residues (indicated by one-letter code) are color-coded in red for intermolecular restraints, black for intramolecular ones, and solid blue for unambiguous restraints (within 0.2 ppm), which could be either of intermolecular or intramolecular nature, respectively. Dashed blue lines restraints with low ambiguity as shown in SI Appendix, Table S2. The 3D structure is represented by the backbone of the two symmetric Aβ(1–42) molecules in one layer color-coded yellow and orange. (B) The distance restraints assigned during the automatic structure calculation by CYANA are displayed on its corresponding 3D structure. Only a single line per residue pair is plotted, even if several restraints exist.
In a second step, an automatic structure calculation was performed by using, in addition to the input data described above, automatically picked peak lists from CHHC, PAR, and PAIN spectra using an iterative assignment process with seven assignment cycles (51). Again, upper distance restraints of 5.5, 7, and 7 Å were implemented for the three spectra, respectively. No lower distance restraints were used. This procedure yielded 551 additional restraints (SI Appendix, Table S3) and resulted in a well-converged structure with an average target function of 1.90 Å2 for the final bundle, comprising the 10 best conformers, and a backbone rmsd of 0.89 for residues 15–42 of the two molecules in the central layer (Fig. 7 and SI Appendix, Table S3). The 3D structure of the automatic calculation superimposes well with the structure from the manual calculation (SI Appendix, Fig. S8). Residue pairs supported by automatically assigned cross-peaks are displayed in Fig. 6B.
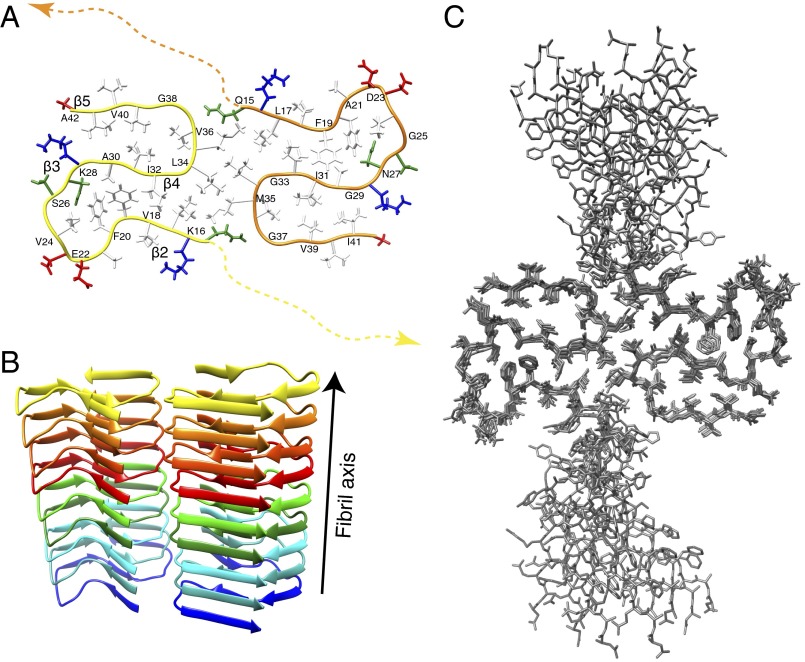
Fig. 7.
The 3D structure of Aβ(1–42) fibrils. (A) Detailed 3D structure of Aβ(1–42) fibrils represented with the conformer showing the smallest CYANA target function. The backbone of the two point symmetric molecules are shown as yellow and orange spines. The 3D structure of the N-terminal residues 1–14 is indicated by dotted lines. The side chains of the positively charged residues are shown in red, the negatively charged in blue, the hydrophobic residues in white, and polar residues in green. Every second residue is labeled with the one-letter amino acid code. (B) A ribbon-based cartoon of the Aβ(1–42) fibrils showing nine molecules of Aβ(1–42) along the fibril axis. Individual molecules are colored following rainbow colors. (C) A bundle of the 10 conformers having an rmsd of 0.89 Å representing the 3D structure is shown.
To double-check the quality of the experimental structure, we calculated the expected peak positions in the CHHC and PAR spectra as expected from the 3D structure. The results are shown in SI Appendix, Figs. S10 and S11, and display excellent agreement. There are virtually no significant peaks not explained by the structure. The fact that some predicted peaks are missing, in particular in the aromatic region, is explained by lower intensity (e.g., to relaxation effects and relayed transfers).
The 3D Structure of Aβ(1–42) Amyloid Fibrils.
The 3D structure of the selected polymorph of Aβ(1–42) amyloid fibrils is composed of two molecules per subunit, which are C2-symmetric with respect to the central axis of the fibril. Each Aβ(1–42) molecule comprises five in-register parallel intermolecular β-strands [i.e., 2–6 (β1), 15–18 (β2), 26–28 (β3), 30–32 (β4), and 39–42 (β5)] (Fig. 4) that wind around two hydrophobic intramolecular cores in a double horseshoe-like manner. The β-sheet β2 thereby interacts through the hydrophobic side chains L17, F19, F20, and V24 with the side-chains A30, I32, and L34 of β-sheet β4. This hydrophobic core is complemented by an asparagine ladder with the side chain of N27 and a glutamine ladder involving the side chain of Q15. Both F19 and F20 face the hydrophobic core requiring a special non–β-strand-like backbone conformation (although the secondary chemical shifts are close to what is expected for a sheet) (Fig. 4), but without having backbone angles in forbidden regions of the Ramachandran plot. Similar arguments are valid for the sequential negatively charged E22 and D23 residues, which are both exposed to the outside. It is interesting to mention that this segment, and in particular the side chains of F19 and F20, are partly stacked off from the main body of its molecule, reaching out to the next layer along the fibril axis. This structural property causes the two fibril ends to be distinct from each other as demonstrated for another polymorph of Aβ(1–42) in an earlier study (18). Note that these four residues are located within the sequence segment F19–K28 that has been identified to be of great significance for the enhanced toxicity of familial mutations in Aβ(1–40) (see also below) and shows structural variability with typically one or two short β-sheets. β-sheet β4 is interacting with β-sheet β5 via the side chains of residues I31 with V36, V39, and I41 on β5, respectively. The intermolecular interaction between the two symmetric Aβ(1–42) molecules involves, in addition to the Q15–M35 and L17–M35 contacts seen in the NMR spectra, hydrophobic contacts between the side chains of L34 and M35. The interaction area between the two monomers in the same layer remains rather small. In contrast to the mainly hydrophobic core, the solvent-exposed surface is composed mostly of polar and charged side chains. Interestingly, a salt bridge is observed between the side chain of K28 and the C terminus of Aβ(1–42), as also recently documented elsewhere (27), but which is very different from what has been observed for the Osaka mutant of Aβ(1–40)ΔE22, where a salt bridge between K28 and the N-terminal E3 is observed (17). Interestingly, the Osaka mutant structure is also the only one described so far with the N-terminal firmly attached and no NMR-invisible residues. As observed in other amyloid structures, the glycine residues are key for the fold. They appear to define the ends of the β-strands (i.e., G25, G29, G37, and G38) (52) and enable the short sharp loops or/and turns by permitting the backbone angles to be in the glycine-specific region of the Ramachandran plot (i.e., G29, G33, and G38). While residues 15–42 are packed densely, the N-terminal segment of residues 1–14 is not entirely rigid. Nonetheless, the presence of a few long-range distance restraints between D1–A42, A2–A42, and F4–V40 indicates that β-sheet β1 is interacting with β-sheet β5 through mainly hydrophobic interactions between the side chains. Such an interaction indicates (Figs. 7 and 8) an intermolecular nature, which might help in reducing the extent of solvent-exposed hydrophobic patches on the fibril.
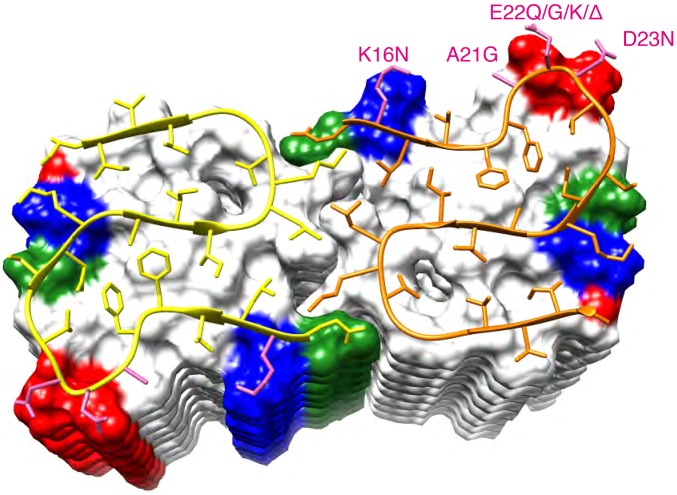
Fig. 8.
Ribbon diagram of the core structure of residues 15–42 of Aβ(1–42) within the fibril. The C2 symmetric molecules are shown as yellow and orange spines, respectively. The β-strands are indicated by arrows. The following molecules within the fibril are shown by a surface representation. Positively and negatively charged surface patches are shown in blue and red, respectively, and all hydrophobic residues in white and polar residues in green. Indicated in pink and labeled accordingly are the residues with known familial Alzheimer’s mutations.
Structure Comparison with the Structural Model of Aβ(1–42) by Ishii and Coworkers (27).
A model for fibrils of Aβ(1–42) has been proposed on the basis of solid-state NMR studies by Ishii and coworkers on the basis of 11 restraints (27). Although in both structures the monomeric units are composed of the double-horseshoe shape as shown in SI Appendix, Fig. S11, a detailed comparison shows large differences within the core structure comprising residues 15–42 with a rmsd of 4.1 Å. Most strikingly is, of course, the number of molecules per fibril layer, which is two in the present study and one in the study by Ishii and coworkers. This is likely due to the absence of MPL information from STEM. Furthermore, there are significant differences in the side-chain packing. For example, in the present 3D structure, the side chain of V36 faces the hydrophobic core composed of residues I31 and V39 (Fig. 7), whereas in the structure by Ishii and coworkers, it faces toward the opposite surface side of their structure (SI Appendix, Fig. S9). Because all of the experimental solid-state NMR-derived distance restraints from the Ishii and coworkers (27) study are satisfied in the structure presented here, which has been determined on many more distance restraints (81 manual and 632 automatic restraints) than the Ishii and coworkers study, the structural differences observed are rather attributed to lack of data there than to real differences.
Discussion
The dimeric double-horseshoe Aβ(1–42) structure presented (Figs. 7 and 8) is of much greater complexity than the peptide amyloid structures determined by X-ray crystallography (25). Although it comprises some of the amyloid-typical structural motifs such as in-register cross–β-sheet secondary structures and Asn/Gln ladders, the complexity of the fold appears to be dictated by burying maximally the hydrophobic side chains—a feature imposed by free energy minimization and also observed in soluble evolved proteins and functional amyloids (53). If maximizing hydrophobic compactness is requested to a sequence that has not been functionally evolved, rather unique structural features may thus arise, such as the two sequential aromatic side chains (i.e., F19 and F20) facing both the hydrophobic core or the two negatively charged side chains (i.e., E22 and D23) facing both the solvent, as well as the now rather well-documented cross–β-sheet motifs and Asn/Gln ladders mentioned above (25). Although not evolutionarily evolved, it is noteworthy to mention that the structure presented here has also been undergoing a selection pressure (albeit a rather primitive one) through three cycles of seeded polymerization starting from a series of polymorphs, which are distinct in the core structure because there are significant differences in the chemical shifts for residues L17, S26, I31, and L34 in the various samples and polymorphs (Fig. 1 and SI Appendix, Fig. S1). Further support for the indication that the presented structure is “fit” in a seeded polymerization are the high similarities of chemical shifts and concomitantly structural similarities with the Aβ(1–42) fibril samples of Ishii and coworkers (27), which also were prepared via seeded polymerization, albeit under different aggregation conditions, while the sample conditions in the work of the Griffin group have been optimized such that no seeding was necessary (40, 54) (SI Appendix, Table S1). The 3D structure of Figs. 7 and 8 may thus represent the conformation of a fast-replicating and fit polymorph. Because replication and cell-to-cell transmissibility are believed to be key mechanisms in AD (55, 56), it is not surprising that the presented Aβ(1–42) polymorph is recognized by the same conformation-specific antibodies as the plaques in AD brain slices (Fig. 2). Thus, the presented 3D structure may be regarded as the culprit of Aβ(1–42) fibril replication in AD. Within this hypothesis, it is interesting to note that many known familial Alzheimer’s mutations that are localized in the core of the presented structure (i.e., K16N, A21G, E22Q, E22G, E22K, and D23N, with the exception of E22Δ) may favor the determined 3D structure (Fig. 8, pink colored side chains): Although most of them may attenuate intermolecular charge repulsion (i.e., K16N, E22Q, E22G, D23N, and E22K), A21G and E22G may instead relax the backbone restraints because of the quite peculiar side-chain arrangements of F19–F20 and E22–E23 (Fig. 7), respectively. In contrast, the further known non-AD-linked single nucleotide polymorphisms within this segment that cause a missense mutation (i.e., V39I, G38V, G38S, G38C, V36M, I32V, I31V, F19L, and V18M) are all conservative mutations (with the exception of G38S and G38C) without expected structural consequences on the Aβ(1–42) fibril structure.
Although we have determined a single disease-relevant Aβ(1–42) fibril structure, still other polymorphs do exist and are probably also the reason for the structural differences between our structure and the models by Grigorieff and coworkers (22) and by Lührs et al. (18) (the latter having an oxidized Met-35).
In summary, the presented 3D structure of a Aβ(1–42) fibril polymorph composed of a complex interplay of hydrophobic interactions and Asn/Gln ladders, which hold together four in register cross–β-strands, may be regarded as a causative agent of Aβ(1–42) fibril replication in AD. It could thus open an avenue for a comprehensive understanding at the atomic level of key aspects of AD, including replication and cell-to-cell transmissibility, as well as structure-assisted developments of potent anti-AD drugs and AD diagnostic markers.
Methods
Protein Expression and Purification.
The plasmid construct containing a N-terminal His-tag followed by a solubility tag (NANP)19 was inserted in Escherichia coli and purified as described (57). The peptide was produced according to the required labeling schemes: uniformly 13C,15N-labeled peptide for the resonance assignment and collection of restraints for the structure calculation, mixed 13C- and 15N-labeled peptide in a ratio of 1:1, and a diluted sample with 13C,15N-labeled monomers diluted in unlabeled ones in a ratio of 1: 3 for the differentiation of intra- and intermolecular restraints.
Fibrillization and Screening of Different Conditions.
The lyophilized material was dissolved with 10 mM NaOH with the help of a sonication bath (three times for 30-s sonication with 50–60% power, interrupted by 1 min cooling on ice). To remove large aggregates, the sample was ultracentrifuged for 1 h at 126,000 × g. The concentration was adjusted as shown in SI Appendix, Table S1. Further phosphate buffer was added to a final concentration of 100 mM at pH of 7.4, and the required additives were added in the sample. Fibrillization was performed under various shaking conditions at 37 °C as indicated in SI Appendix, Table S1. Seeding was performed for three generations, with 10% of the grandparent generation used as seeds for the parent generation, and again 10% for the daughter generation.
Filling the Rotor and Solid-State NMR Measurements.
After the last seeding step, the fibrils, made up of 15–20 mg of peptide, were centrifuged at 30,000 × g overnight (SW41-TI swinging bucket, optima L90-K; Beckman) and resuspended in MilliQ water. The fibrils were washed for 3 d by gentle shaking. The pellet was again centrifuged at 30,000 × g overnight, the supernatant was discarded, and the fibrils were packed into a 3.2-mm Bruker rotor by ultracentrifugation using a homemade filling device (58). The drive tip was sealed with epoxy glue (Araldite blue) to prevent dehydration of the sample.
The 2D NCA and DARR experiments, for sample screening (SI Appendix, Fig. S1), were measured on a Bruker AVANCE III 600-MHz spectrometer using a 3.2-mm triple-resonance probe. The 2D DARR, CHHC, PAR, PAIN, NHHC, and TEDOR spectra measured to derive restraints were obtained on a Bruker AVANCE III 850 MHz using a 3.2-mm triple-resonance probe. Experimental details are listed in SI Appendix, Tables S4 and S5. The sample temperature was adjusted to 4 °C by using the water resonance of supernatant water (58). Data were processed with Topspin (v3.1) and analyzed with CcpNmr Analysis (v2.3) (59).
MPL Measurement.
Two samples with an Aβ(1–42) concentration of 100 μg/mL were prepared for the MPL analysis. The fibrils in solution were attached to the carbon substrate and washed rigorously to remove residual salts. After washing, the specimen was freeze-dried. The measurements were performed by using different concentrations of the fibrils (no, 3×, and 10× dilution). The analysis of the data was performed with the software PCMass32 (available from the Brookhaven STEM resource openwetware.org/wiki/STEM_Facility). For the analysis established models were used as a starting point and tuned for the respective Aβ(1–42) fibrils. The radius of integration was chosen between 80 and 120 Å according to the width of the filaments. Only well-isolated filaments with a background rmsd > 0.5 were chosen. To control for possible artifacts induced by the presence of detergents or salts, tobacco mosaic virus was codeposited as an internal mass calibration standard on all specimens, and its MPL value was monitored in all specimens. The MPL analysis of the tobacco mosaic virus showed a value of 13.1 kDa/Å with a SD of 2% as expected.
Dot Blot Analysis.
Dot blot assays were conducted to detect the immunoreactivity of Altspective (no, 3×, and 10× dilution). Previous publications describe the production of the study of Altspective (no, 3×, and 10× αAPFn) (60), A11 (61), and OC (35). Kayed et al. (34) and Hatami et al. (36) describe the methods for generating the A11- and OC-like monoclonal antibodies used in the study, respectively. The 6E10 and 4G8 antibodies are commonly used mouse antibodies for Aβ from Covance. Aβ(1–42) fibril samples were spotted on a nitrocellulose membrane at ∼3 μg per sample. Additionally, three samples of different Aβ conformations—Aβ(1–40) oligomer mimics (0.1 μg per spot), Aβ(1–42) fibrils (standard from the C.G.G. laboratory) (0.3 μg per spot), and Aβ(1–40) monomers (1 μg per spot)—were applied to the membrane as the controls for different antibodies. The membranes with the samples were blocked in 10% (vol/vol) nonfat dry milk in Tris-buffered saline containing 0.01% Tween 20 (TBS-T) at room temperature for 1 h. After three repetitions of 5-min TBS-T washes, the membranes were probed with αAβ(4G8, 6E10 at 1:10,000) or conformation-specific antibodies [A11, 1:500; αAPF; 1:5,000 OC, 1:10,000; mA55 (1:1,000), mA118, m204 (1:2,000), mA204 (1:2,000), mOC hybridoma supernatants (1:100)] overnight at 4 °C. The membranes were washed three times and then incubated with either anti-mouse (4G8 and 6E10) or anti-rabbit (all others) IgG conjugated with horseradish peroxidase (1:10,000; Jackson Laboratory) at room temperature for 1 h. The blots were then developed with the ECL chemiluminescence kit from ThermoFisher Scientific (SuperSignal West Pico). The results of the dot blots were imaged by using a Nikon D700 camera as described (62). Fig. 2A was uniformly digitally brightened twofold.
Immunohistochemistry.
Postmortem paraformaldehyde-fixed brain tissue was obtained under an Institutional Review Board-approved protocol from the neuropathological core of the University of California at Irvine Alzheimer’s Disease Research Center. Broadmann’s area 11 of the frontal cortex from case no. 07-03 (plaque stage A) was examined by immunohistochemistry as described in detail (36). The 40-µm-thick sections were incubated with 1 µg/mL primary antibody for 12 h at 4 °C, washed, and incubated with biotinylated goat anti-rabbit or biotinylated horse anti-mouse IgG (Vector Laboratories) for 1 h at 25 °C. After incubation with the secondary antibodies, the tissue sections were washed, and an ABC peroxidase kit and 3,3′-diaminobenzidine substrate kit (Vector Laboratories) were used to detect the biotinylated secondary antibodies.
Limited Proteolysis.
Trypsin and proteinase K were dissolved in 50 mM Tris⋅HCl (pH 8.0) at a concentration of 1 mg/mL and always kept on ice. The preformed Aβ(1–42) fibrils (see above) were added to the protease with an approximate protease:Aβ(1–42) ratio of 1:10 (wt/wt). (It was not possible to measure the mass of the fibrils accurately, because of the low concentration of 30 μM peptide used for the fibrillization and the presence of only one tyrosine residue in the sequence, which leads to a somewhat inaccurate UV-Vis A280 value.) The digestion was performed at 37 °C under gentle shaking. The reaction was quenched by adding 2 mM phenylmethylsulfonylfluorid after different time points from 30 min to overnight. To each time point, a control was prepared with the addition of the same amount of buffer without the protease. After digestion, the fibrils were centrifuged at 126,000 × g for 15 min and dissolved in a 85% DMSO solvent with 5% D2O, 0.1% TFA, and 10% H2O. [15N,1H]-TROSY spectra of the dissolved fibrils were recorded at 298 K on a Bruker 700 MHz Avance III spectrometer equipped with a triple-resonance cryoprobe (SI Appendix, Fig. S5). Peak intensities of the individual 15N-1H-moieties were normalized relative to the corresponding spectrum of the control sample without digestion.
Structure Calculation.
Structure calculations followed the protocol established for fibril structure determinations by solid-state NMR with CYANA (17, 63). Structure calculations were performed for a fibril comprising three layers with two molecules per layer. The six monomers were held in identical conformation by dihedral angle difference restraints for all corresponding torsion angles, and a symmetric relative orientation of the monomers was maintained by distance difference restraints between symmetry-related intermolecular Cα–Cα distances (64). The five parallel β-sheets were restrained by upper and lower bound distance restraints of 1.8 ≤ dOH ≤ 2.0 Å and 2.7 ≤ dON ≤ 3.0 Å. For each possible combination of the directions of the five β-sheets (up or down along the fibril axis; 24 = 16 possibilities), a separate calculation was performed. Backbone torsion angle restraints were derived from chemical shifts as discussed in The 3D Structure Determination of Aβ(1–42) Amyloid Fibrils (50). Before the structure calculation, the restraints were replicated according to the fibril symmetry (e.g., six times for intramolecular restraints, three times for intermolecular restraints between the two molecules in one fibril layer, and four times for the hydrogen-bond restraints connecting two molecules in adjacent layers). A first structure calculation was performed by using the 81 manually assigned distance restraints listed in SI Appendix, Table S2, which are either spectrally unambiguous (10 restraints) or of low spectral ambiguity. A total of 20 restraints were unambiguously intramolecular, and 3 were unambiguously intermolecular. Starting from random torsion angle values, 500 conformers were calculated by using 35,000 torsion angle dynamics steps in CYANA, and the 10 conformers with lowest target function values were retained for analysis. Only one possible combination of the directions of the five β-sheets yielded a low target function, and we continued with this one only. This structure was subsequently used as additional input for a structure calculation with automated distance restraint assignment (51). The first cycle of combined automated distance restraint assignment and structure calculation was skipped and replaced by the aforementioned structure. Chemical shift tolerances of 0.3 ppm for 13C and 0.6 ppm for 15N were used for the automated distance restraint assignment. Hydrogen-bond distance restraints and manually assigned NMR distance restraints were applied with weights of 10 and 3, respectively, relative to those for automatically assigned distance restraints. The final structure was calculated as a consensus structure bundle (65) of 10 conformers that represent the 3D structure of Aβ(1–42) fibrils. Structural statistics are given in SI Appendix, Table S3. The chemical-shift assignment of Aβ(1–42) fibrils has been deposited in the BMRB (accession no. 26692) and the 3D structure in the Protein Data Bank (PDB ID code 2NAO).
Note added in proof.
After this manuscript was accepted we learned of another paper [Colvin MT, et al. (2016) J Am Chem Soc 137(23):7509–7518] describing the structure of Aβ1–42 fibrils with a Met at position 0. The two samples have a very similar chemical-shift fingerprint and the structure of the highly ordered region (residues 15–42) is virtually identical.
Supplementary Material
Acknowledgments
We thank Beth Y. Lin (Brookhaven National Laboratory) for the STEM measurements; Frank Delaglio for support with NMRPipe; and Peter Tittmann (Scientific Center for Optical and Electron Microscopy of ETH) for support with electron microscopy. This work was supported by Swiss National Science Foundation Grants 200020_159707 and 200020_146757; and by French Agence Nationale de la Recherche Grants ANR-11-BSV8-021-01, 0013-01, and ANR-14-CE09-0024B.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2NAO), and the Biological Magnetic Resonance Bank, www.bmrb.wisc.edu/ (accession no. 26692).
See Commentary on page 9398.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600749113/-/DCSupplemental.
References
- 1.Masters CL, et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82(12):4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW. Alzheimer’s disease and Down’s syndrome: Sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122(3):1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 3.Kang J, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325(6106):733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Costanzo M, Zurzolo C. The cell biology of prion-like spread of protein aggregates: Mechanisms and implication in neurodegeneration. Biochem J. 2013;452(1):1–17. doi: 10.1042/BJ20121898. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roher AE, et al. beta-Amyloid-(1-42) is a major component of cerebrovascular amyloid deposits: Implications for the pathology of Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(22):10836–10840. doi: 10.1073/pnas.90.22.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selkoe DJ. Alzheimer’s disease: A central role for amyloid. J Neuropathol Exp Neurol. 1994;53(5):438–447. doi: 10.1097/00005072-199409000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Iwatsubo T, et al. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: Evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13(1):45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 9.Gravina SA, et al. Amyloid beta protein (A beta) in Alzheimer’s disease brain. Biochemical and immunocytochemical analysis with antibodies specific for forms ending at A beta 40 or A beta 42(43) J Biol Chem. 1995;270(13):7013–7016. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- 10.Meier BH, Böckmann A. The structure of fibrils from ‘misfolded’ proteins. Curr Opin Struct Biol. 2015;30:43–49. doi: 10.1016/j.sbi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32(18):4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 12.Jarrett JT, Berger EP, Lansbury PT., Jr The C-terminus of the beta protein is critical in amyloidogenesis. Ann N Y Acad Sci. 1993;695:144–148. doi: 10.1111/j.1749-6632.1993.tb23043.x. [DOI] [PubMed] [Google Scholar]
- 13.El-Agnaf OM, Mahil DS, Patel BP, Austen BM. Oligomerization and toxicity of beta-amyloid-42 implicated in Alzheimer’s disease. Biochem Biophys Res Commun. 2000;273(3):1003–1007. doi: 10.1006/bbrc.2000.3051. [DOI] [PubMed] [Google Scholar]
- 14.Bertini I, Gonnelli L, Luchinat C, Mao J, Nesi A. A new structural model of Aβ40 fibrils. J Am Chem Soc. 2011;133(40):16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- 15.Petkova AT, et al. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99(26):16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schütz AK, et al. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl. 2015;54(1):331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lührs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide solvent protection analysis demonstrates that amyloid-beta(1-40) and amyloid-beta(1-42) form different fibrillar structures under identical conditions. Biochem J. 2007;404(1):63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirschner DA, Abraham C, Selkoe DJ. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc Natl Acad Sci USA. 1986;83(2):503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273(3):729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt M, et al. Peptide dimer structure in an Aβ(1-42) fibril visualized with cryo-EM. Proc Natl Acad Sci USA. 2015;112(38):11858–11863. doi: 10.1073/pnas.1503455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed M, et al. Structural conversion of neurotoxic amyloid-β(1-42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17(5):561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto A, et al. Analysis of the secondary structure of beta-amyloid (Abeta42) fibrils by systematic proline replacement. J Biol Chem. 2004;279(50):52781–52788. doi: 10.1074/jbc.M406262200. [DOI] [PubMed] [Google Scholar]
- 25.Nelson R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435(7043):773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41(51):15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Y, et al. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22(6):499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fändrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion. 2009;3(2):89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colletier J-P, et al. Molecular basis for amyloid-beta polymorphism. Proc Natl Acad Sci USA. 2011;108(41):16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bousset L, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gath J, et al. Unlike twins: An NMR comparison of two α-synuclein polymorphs featuring different toxicity. PLoS One. 2014;9(3):e90659. doi: 10.1371/journal.pone.0090659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takegoshi K, Nakamura S, Terao T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344(5-6):631–637. [Google Scholar]
- 33.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-driven 13C-13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118(5):2325–2341. [Google Scholar]
- 34.Kayed R, et al. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar Aβ oligomers. Mol Neurodegener. 2010;5(1):57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayed R, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2(1):18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatami A, Albay R, 3rd, Monjazeb S, Milton S, Glabe C. Monoclonal antibodies against Aβ42 fibrils distinguish multiple aggregation state polymorphisms in vitro and in Alzheimer disease brain. J Biol Chem. 2014;289(46):32131–32143. doi: 10.1074/jbc.M114.594846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller SA, Engel A. Structure and mass analysis by scanning transmission electron microscopy. Micron. 2001;32(1):21–31. doi: 10.1016/s0968-4328(00)00022-6. [DOI] [PubMed] [Google Scholar]
- 38.Sousa AA, Leapman RD. Mass mapping of amyloid fibrils in the electron microscope using STEM imaging. Methods Mol Biol. 2013;950(Chapter 12):195–207. doi: 10.1007/978-1-62703-137-0_12. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D, Schultz P, Steven AC, Wall JS. Mass analysis of biological macromolecular complexes by STEM. Biol Cell. 1994;80(2-3):181–192. doi: 10.1111/j.1768-322x.1994.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 40.Ravotti F, et al. Solid-state NMR sequential assignment of an Amyloid-β(1-42) fibril polymorph. Biomol NMR Assign. May 10, 2016 doi: 10.1007/s12104-016-9682-y. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewandowski JR, De Paëpe G, Griffin RG. Proton assisted insensitive nuclei cross polarization. J Am Chem Soc. 2007;129(4):728–729. doi: 10.1021/ja0650394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J Am Chem Soc. 2002;124(36):10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 44.Hing A, Vega S, Schaefer J. Transferred-echo double-resonance NMR. J Magn Reson. 1992;96(1):205. [Google Scholar]
- 45.Luginbuhl P, Szyperski T, Wüthrich K. Statistical basis for the use of 13 C α chemical shifts in protein structure determination. J Magn Reson B. 1995;109:229–233. [Google Scholar]
- 46.Wishart DS, Sykes BD. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4(2):171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 47.Wishart DS, Sykes BD, Richards FM. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31(6):1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 48.Lange A, Luca S, Baldus M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J Am Chem Soc. 2002;124(33):9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 49.De Paëpe G, Lewandowski JR, Loquet A, Böckmann A, Griffin RG. Proton assisted recoupling and protein structure determination. J Chem Phys. 2008;129(24):245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 51.Güntert P, Buchner L. Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR. 2015;62(4):453–471. doi: 10.1007/s10858-015-9924-9. [DOI] [PubMed] [Google Scholar]
- 52.Daskalov A, et al. Contribution of specific residues of the β-solenoid fold to HET-s prion function, amyloid structure and stability. PLoS Pathog. 2014;10(6):e1004158. doi: 10.1371/journal.ppat.1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 54.Colvin MT, et al. High resolution structural characterization of Aβ42 amyloid fibrils by magic angle spinning NMR. J Am Chem Soc. 2015;137(23):7509–7518. doi: 10.1021/jacs.5b03997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riek R. Cell biology: Infectious Alzheimer’s disease? Nature. 2006;444(7118):429–431. doi: 10.1038/444429a. [DOI] [PubMed] [Google Scholar]
- 56.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313(5794):1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 57.Wälti MA, Orts J, Vögeli B, Campioni S, Riek R. Solution NMR studies of recombinant Aβ(1-42): From the presence of a micellar entity to residual β-sheet structure in the soluble species. ChemBioChem. 2015;16(4):659–669. doi: 10.1002/cbic.201402595. [DOI] [PubMed] [Google Scholar]
- 58.Böckmann A, et al. Characterization of different water pools in solid-state NMR protein samples. J Biomol NMR. 2009;45(3):319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 59.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 60.Kayed R, et al. Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J Biol Chem. 2009;284(7):4230–4237. doi: 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 62.Khoury MK, Parker I, Aswad DW. Acquisition of chemiluminescent signals from immunoblots with a digital single-lens reflex camera. Anal Biochem. 2010;397(1):129–131. doi: 10.1016/j.ab.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Melckebeke H, et al. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132(39):13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 64.Lin YJ, Kirchner DK, Güntert P. Influence of ¹H chemical shift assignments of the interface residues on structure determinations of homodimeric proteins. J Magn Reson. 2012;222:96–104. doi: 10.1016/j.jmr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Buchner L, Güntert P. Increased reliability of nuclear magnetic resonance protein structures by consensus structure bundles. Structure. 2015;23(2):425–434. doi: 10.1016/j.str.2014.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.