Significance
Bacillus thuringiensis and its toxins are widely used for insect control. Notwithstanding the remarkable importance of this insect pathogen, its killing mechanism has yet to be fully elucidated. Here we show that the microbiota resident in the host midgut triggers a lethal septicemia. The infection process is enhanced by reducing the host immune response and its control on replication of midgut bacteria invading the body cavity through toxin-induced epithelial lesions. The experimental approach used, leaving the midgut microbiota unaltered, allows identification of the bacterial species switching from resident symbionts to pathogens and sets the stage for developing new insect biocontrol technologies based on host immunosuppression as a strategy to enhance the impact of natural antagonists.
Keywords: bioinsecticide, insect–pathogen interactions, insect biocontrol, pore-forming toxins, immunity
Abstract
Bacillus thuringiensis is a widely used bacterial entomopathogen producing insecticidal toxins, some of which are expressed in insect-resistant transgenic crops. Surprisingly, the killing mechanism of B. thuringiensis remains controversial. In particular, the importance of the septicemia induced by the host midgut microbiota is still debated as a result of the lack of experimental evidence obtained without drastic manipulation of the midgut and its content. Here this key issue is addressed by RNAi-mediated silencing of an immune gene in a lepidopteran host Spodoptera littoralis, leaving the midgut microbiota unaltered. The resulting cellular immunosuppression was characterized by a reduced nodulation response, which was associated with a significant enhancement of host larvae mortality triggered by B. thuringiensis and a Cry toxin. This was determined by an uncontrolled proliferation of midgut bacteria, after entering the body cavity through toxin-induced epithelial lesions. Consequently, the hemolymphatic microbiota dramatically changed upon treatment with Cry1Ca toxin, showing a remarkable predominance of Serratia and Clostridium species, which switched from asymptomatic gut symbionts to hemocoelic pathogens. These experimental results demonstrate the important contribution of host enteric flora in B. thuringiensis-killing activity and provide a sound foundation for developing new insect control strategies aimed at enhancing the impact of biocontrol agents by reducing the immunocompetence of the host.
The entomopathogenic Gram-positive bacterium, Bacillus thuringiensis, produces parasporal crystalline inclusions containing proteinaceous toxins (δ-endotoxins, Cry and Cyt) during sporulation and secretes a different set of insecticidal proteins during its vegetative growth stage (1–4). When ingested by susceptible insects, the δ-endotoxins are processed by gut proteases and give rise to active fragments able to bind protein receptors located on the brush border of midgut cells, where, upon oligomerization, they form cation-permeable pores, triggering the osmotic lysis of cells and subsequent death of the insect (3, 5, 6) (SI Appendix, Fig. S1).
The discovery of B. thuringiensis and the remarkable interest generated by its selective insecticidal activity have triggered a number of investigations aimed at developing successful biocontrol applications and bioinspired insect control technologies. B. thuringiensis spores and crystals have been successfully used for spray applications against insect pests and pathogen vectors (7, 8). Moreover, the isolation of the genes coding for B. thuringiensis toxins has allowed the development of transgenic B. thuringiensis crops, a pioneering biotechnology for insect control in agriculture that reached the market in 1996 and has been successfully adopted in many regions around the world (9). Transgenic B. thuringiensis plant technology has been greatly improved to confer protection against different pests and to limit the selection of resistant strains through gene pyramiding (i.e., the coexpression of different B. thuringiensis toxins in a plant) (10–12).
The management of insect resistance has attracted considerable attention as a key issue for effectively deploying this bioinsecticide (13–16). Indeed, field monitoring of pest susceptibility, isolation of new toxins, and the study of resistance mechanisms have been widely pursued by the scientific community (10, 15). The growing evidence that the alteration of toxin binding is the most common resistance mechanism (6) emphasizes the importance of binding models to predict the effective toxin combinations for successful implementation of gene pyramiding strategies (12, 16–19).
Comparatively less attention has been devoted to other important research aspects, such as the complex network of molecular interactions underpinning the host killing mechanism and the role of factors other than the pore-forming toxins that contribute to B. thuringiensis pathogenicity and virulence (20, 21). A heated debate on the B. thuringiensis killing mechanism started almost a decade ago, and a consensus has not yet been reached. Indeed, divergent opinions persist on the role played by the microbiota residing in the midgut of lepidopteran hosts and the septicemia it may cause after toxin-induced disruption of the lining epithelium (22–31). Whether the gut paralysis and associated feeding cessation observed in larvae exposed to Cry pore-forming toxins or the septicemia induced by bacterial proliferation in the insect body cavity is the major factor contributing to eventual death remains to be resolved (5, 32) (SI Appendix, Fig. S1). The various experimental approaches adopted to address this important research question have led to different conclusions that are not easy to reconcile. Therefore, whether B. thuringiensis is a virulent pathogen that disrupts gut functionality or requires the cooperative action of the commensal midgut microbiota to kill the host remains elusive.
It is important to narrow this research gap by defining the effective role of the host microbial community in the induction of B. thuringiensis toxin’s adverse effects. This will significantly influence the development of new insect control strategies based on the manipulation of the complex immune interplay between insect hosts and their midgut microbiota, which appears to be far more complex and important than previously thought (33).
Here we have addressed this relevant research issue using an experimental approach that provides insight into the importance of septicemia in the complex killing mechanism mediated by B. thuringiensis toxins. To circumvent the problems associated with the manipulation of the midgut microbiota with antibiotics (22–25, 27–31) or of the immune response with immunosuppressive molecules (26, 34), both of which have unpredictable multiple effects, we performed selective RNAi-mediated silencing of an immune gene (102 Sl, 102 Spodoptera littoralis), recently identified in Spodoptera littoralis (35). This gene is highly expressed in the immune cells circulating in the hemolymph (i.e., hemocytes) and modulates their incorporation in multilayered capsules formed around foreign intruders entering the body cavity (i.e., the hemocoel) (35). The final result is the encapsulation of large objects and the nodulation of bacterial cells or aggregates, which are eventually suppressed by localized production of melanin and other toxic compounds on their surface (36, 37).
In the present work, we used the RNAi-mediated disruption of the cellular immune barriers for an unbiased characterization of the changes in hemolymph microbiota promoted by B. thuringiensis toxin-induced lesions and assessment of associated mortality. The results clearly indicate the important role of septicemia in the induction of host death, and set the stage to develop innovative insect control strategies that may enhance the impact of biocontrol agents by altering the immunocompetence of the host.
Results
Effect of 102 Sl Silencing on Host Immune Response.
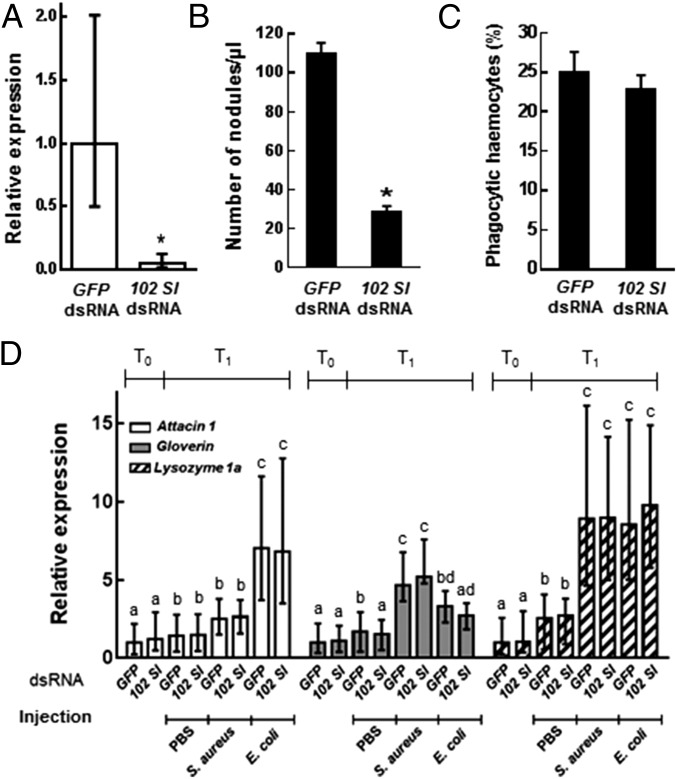
Previous work has shown that 102 Sl is directly involved in the regulation of encapsulation and melanization reactions by S. littoralis larvae (35). Because nodulation is an important antimicrobial immune response that shows functional similarity to the encapsulation of large foreign intruders (36), it was reasonable to investigate whether silencing of the 102 Sl gene in S. littoralis larvae has any effect on nodulation and, as a consequence, on the proliferation of microbial pathogens. We found that indeed this is the case; in larvae orally treated with 102 Sl dsRNA and subsequently challenged by hemocoelic injection of bacterial cells (Escherichia coli), the silencing of the target gene (P < 0.0001, t = 18.98, df = 34, n = 18) (Fig. 1A) was associated with a significant reduction in nodule formation (P < 0.0001, t = 26.0, df = 54, n = 28) (Fig. 1B).
Fig. 1.
Cellular and humoral immune response by S. littoralis larvae as affected by RNAi-mediated silencing of the immune gene 102 Sl. (A) The transcript level of 102 Sl was significantly down-regulated in S. littoralis larvae by oral administration of 102 Sl dsRNA (P < 0.0001, t = 18.98, df = 34, n = 18). (B) RNAi-mediated silencing of 102 Sl significantly reduced the number of nodules present in the hemolymph of S. littoralis larvae that received an injection of E. coli cells (P < 0.0001, t = 25.994, df = 27, n = 28). (C) In contrast, phagocytosis of fluorescein-conjugated E. coli cells was not influenced by gene silencing. (D) The transcript level of genes encoding the humoral effectors considered was significantly enhanced by the immune challenge (n = 8 for each sampling point; attacin 1: F3, 66 = 198.13, P < 0.0001; gloverin: F3, 66 = 39.58, P < 0.0001; lysozyme1a: F3, 66 = 95.60, P < 0.0001), but was not influenced by gene silencing. The values reported are the mean ± SE. T0 is the time of injection, T1 is 18 h after injection. Different letters denote significant differences between treatments compared within each gene considered.
To assess whether the 102 Sl silencing had any impact on other components of the immune response, we analyzed its effect on the phagocytic activity of hemocytes and on the transcription profiles of genes encoding humoral effectors, including antimicrobial peptides (AMPs) and lysozyme (38, 39), following an immune challenge. The internalization of FITC-labeled E. coli by hemocytes was not affected by gene silencing (Fig. 1C). The injection of bacteria significantly enhanced transcription in the hemocytes of genes encoding these selected humoral effectors (attacin 1: F3, 66 = 198.13, P < 0.0001; gloverin: F3, 66 = 39.58, P < 0.0001; lysozyme 1a: F3, 66 = 95.60, P < 0.0001), but this was not influenced by gene silencing (Fig. 1D and SI Appendix, Table S1). The same response pattern was observed in the midgut and fat body (SI Appendix, Fig. S2 and Table S1).
Susceptibility of Immunosuppressed S. littoralis Larvae to B. thuringiensis and Cry1Ca.
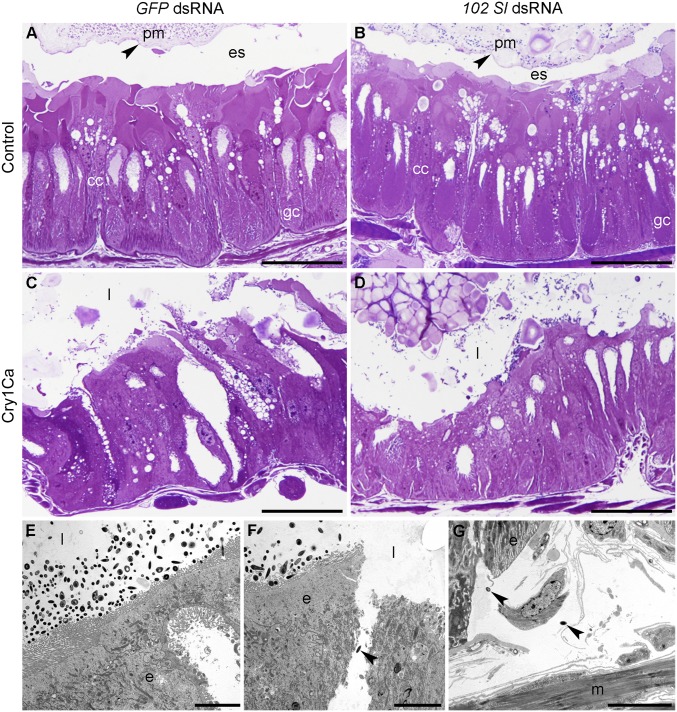
Histological analysis showed striking alterations of the peritrophic matrix (PM) and midgut epithelium in larvae that ingested Cry1Ca, irrespective of previous dsRNA treatment received (Fig. 2 A–D). Disruption of the PM coincided with the cessation of feeding upon intoxication and was reproduced by forced starvation, suggesting that feeding cessation upon intoxication is likely responsible for this alteration (SI Appendix, Fig. S3). The PM disaggregation allowed direct contact of bacteria with the apical membrane of the midgut epithelium, which had lost its integrity as a consequence of the toxin-induced lesions. Transmission electron microscopy observations of larvae exposed to Cry1Ca showed a massive presence of bacteria adjacent to the apical brush border of midgut epithelial cells (Fig. 2E). Some of these bacteria had entered the hemocoel through damaged areas of the gut barrier (Fig. 2 F and G).
Fig. 2.
Midgut morphology of experimental S. littoralis larvae. (A and B) Semithin cross-sections of the midgut epithelium from control larvae treated with GFP dsRNA (A) or 102 Sl dsRNA (B). The epithelial monolayer is intact, showing regular columnar cells (cc) and goblet cells (gc) and evident PM (pm; arrowhead), delimiting the ectoperitrophic space (es). (C and D) Semithin cross-sections of the midgut epithelium from control larvae treated with GFP dsRNA (C) or 102 Sl dsRNA (D) and fed with Cry1Ca. The toxin induces severe alterations in both samples and disrupts the integrity of the intestinal barrier. Alterations of cell morphology are visible. The disaggregation of the PM, associated with feeding cessation, is evident (see the text for details), and the midgut lumen (l) does not show the presence of a delimited ectoperitrophic space. (E–G) Transmission electron microscopy analysis of the midgut epithelium (e) explanted from experimental larvae treated with Cry1Ca shows signs of cellular alteration (E–G) and the presence of bacterial cells close to the apical brush border of the midgut (E), with some of them located in the disrupted areas of the lining epithelium (arrowhead; F) and in the hemocoel (arrowhead), where muscle fibers (m) are evident (G). (Scale bars: 50 μm in A–D, 5 μm in E–G.)
We reasoned that the bacterial invasion of the body cavity following B. thuringiensis toxin ingestion could be modulated by the immunocompetence of the host larvae. Thus we investigated whether the negative impact on nodulation exerted by 102 Sl silencing might have a synergistic interaction with exposure to B. thuringiensis or its toxins. We found that indeed this was the case. The susceptibility of S. littoralis larvae treated with 102 Sl dsRNA was significantly greater than that observed in controls treated with GFP dsRNA, both for the commercial formulation of B. thuringiensis (Xentari) and the purified toxin (Cry1Ca) (Table 1 and SI Appendix, Fig. S4). On administration of 102 Sl dsRNA, an approximate fivefold increase in mortality was recorded for both treatments (Table 1 and SI Appendix, Fig. S4). Exposure to any dsRNA in the absence of B. thuringiensis or its toxin was associated with a survival rate nearly identical to that observed for untreated controls (91%; n = 154); these experimental larvae all underwent pupation and developed into adults.
Table 1.
Enhancement of B. thuringiensis toxicity by host immunosuppression
| LC50† | |||
| Treatment* | GFP dsRNA | 102 Sl dsRNA | TI‡ |
| Xentari | 36.6 (20.2–64.4) | 7.1 (3.9–13.0) | 5.1 (2.3–11.3) |
| Cry1Ca | 6.6 (2.8–16.9) | 1.2 (0.5–2.5) | 5.5 (1.9–16) |
Toxicity of Xentari and Cry1Ca toxin was obtained on S. littoralis larvae after RNAi-mediated silencing of an immune gene (102 Sl dsRNA), and in immunocompetent controls (GFP dsRNA).
Concentration (micrograms of toxin or Xentari per square centimeter of diet) killing 50% of experimental larvae, with 95% fiducial limits reported in parentheses.
The toxicity increase (TI) is calculated as the ratio between LC50 values scored in GFP dsRNA control larvae and in larvae treated with 102 Sl dsRNA; 95% fiducial limits are reported in parentheses.
Collectively, these data suggest that the toxin-induced lesions allow the entrance of midgut bacteria into the hemocoel, and that the diminished immunocompetence induced by 102 Sl silencing has a major impact on the resulting levels of infection, which could account for a significant part of the B. thuringiensis killing mechanism.
Septicemia Induced by Cry1Ca Treatment.
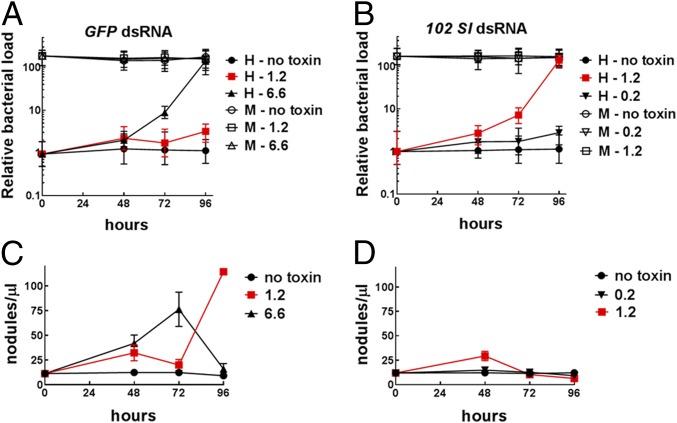
To test the hypothesis that septicemia plays a key role in B. thuringiensis toxin-induced mortality, we analyzed the impact of different toxin doses and immunosuppression on the bacterial load in the hemolymph. The numbers of bacteria recovered from hemolymph samples increased significantly over time (F3, 176 = 310.36, P < 0.0001), was directly correlated with toxin dose (F3, 176 = 45.65, P < 0.0001), and was significantly influenced by gene silencing (F1, 176 = 22.65, P < 0.0001) (Fig. 3 A and B). The different LC50 values observed in control and silenced larvae (6.6 and 1.2 µg/cm2, respectively) (Table 1 and SI Appendix, Fig. S4) were associated with similar bacterial loads in the hemolymph (Fig. 3 A and B and SI Appendix, Table S2). Conversely, the same dose of toxin (1.2 µg/cm2) resulted in an exponential growth of bacteria and lethal septicemia only in silenced experimental larvae (Fig. 3 A and B, red curves). In all cases, the lethal bacterial proliferation (Fig. 3 A and B) was associated with failure of the nodulation response (Fig. 3 C and D), which was significantly influenced by both toxin exposure (F2, 81 = 16.94, P < 0.0001) and gene silencing (F1, 81 = 30.63, P < 0.0001). Collectively, these data clearly demonstrate significantly impaired immunocompetence by gene silencing that favored bacterial replication into the hemolymph and enhanced larval mortality in the experimental larvae exposed to effective doses of B. thuringiensis toxin.
Fig. 3.
Relative quantification of bacterial load by qRT-PCR and nodulation response. (A and B) Fold-change over time of bacterial load in S. littoralis larvae, as affected by different doses (µg/cm2) of Cry1Ca, in controls exposed to GFP dsRNA (A) and in immunosuppressed individuals that received a 102 Sl dsRNA treatment (B). The bacterial load resulted significantly influenced by all experimental factors only in the hemolymph (H) environment (dsRNA: F1, 176 = 22.65, P < 0.0001; Cry1Ca: F3, 176 = 45.65, P < 0.0001; time: F3, 176 = 310.36, P < 0.0001), whereas no significant changes were observed in the midgut (M). (C and D) The concurrent nodulation response in controls exposed to GFP dsRNA (C) and in immunosuppressed individuals who received 102 Sl dsRNA treatment (D) clearly evidenced an inverse correlation between bacterial growth and nodulation, which was significantly influenced by both toxin exposure (F2, 81 = 16.94, P < 0.0001) and gene silencing (F1, 81 = 30.63, P < 0.0001).
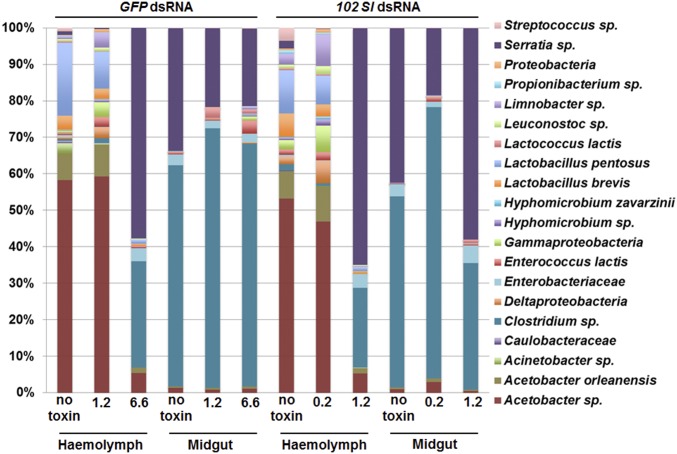
The 16S rRNA amplicon-based analysis of the microbiota of the midgut and hemocoel clearly indicate that B. thuringiensis-induced lesions allow the passage of bacteria through the intestinal barrier and subsequent colonization of the body cavity (Fig. 4). The bacterial profile of the midgut and hemolymph were very different in untreated controls, and became similar only in larvae exposed to an LC50 dose of B. thuringiensis toxin (6.6 µg/cm2 for GFP dsRNA-treated larvae and 1.2 µg/cm2 for 102 Sl dsRNA-treated larvae) (Fig. 4), which caused septicemia in the experimental larvae (Fig. 3 A and B). These results are independently supported by a phylogenetic beta diversity analysis showing a clear separation between hemolymph samples from untreated larvae and samples of midgut and hemolymph obtained from larvae exposed to an LC50 toxin dose (SI Appendix, Fig. S5). Alpha diversity measures indicated a reduction of microbial diversity in the hemolymph on treatment with higher doses of toxin (SI Appendix, Table S3). The low numbers of bacteria in the hemolymph of untreated larvae included species mainly of the genera Acetobacter, Enterococcus, and Lactobacillus, whereas the structure of the hemolymphatic microbiota changed dramatically on treatment with an LC50 dose, resulting in almost exclusive colonization by Serratia and Clostridium, characteristic of the midgut environment (Fig. 4 and SI Appendix, Table S4).
Fig. 4.
Incidence of the major bacterial taxonomic groups detected by pyrosequencing. Relative abundance of identified microbial taxa in the midgut content and hemolymph samples collected from S. littoralis larvae treated with 102 Sl dsRNA or GFP dsRNA, and exposed to various doses (µg/cm2) of Cry1Ca. Sample IDs are listed in SI Appendix, Table S4.
Collectively, these results demonstrate that the septicemia caused by midgut bacteria and induced by B. thuringiensis toxins even in the absence of bacterial spores is responsible for host mortality, which is more pronounced when immunocompetence is reduced.
Discussion
The immune response of insects has tightly linked humoral and cellular components that provide protection against microbial pathogens and metazoan parasites (39). We have recently discovered that in Lepidoptera, regulation of the encapsulation and melanization process is mediated in part by a molecular scaffold composed of amyloid fibers (37), and that the RNAi-mediated silencing of the gene encoding the amyloidogenic protein (102 Sl) disrupts these immune responses (35). Here we have demonstrated that in S. littoralis, the nodulation/suppression of microbes is similarly affected by the silencing of this gene, whereas the phagocytic activity of hemocytes and the humoral antimicrobial response remain unaltered.
This targeted immunosuppression results in enhanced host sensitivity to microbial entomopathogens, paving the way for an unbiased assessment of the role played by septicemia in the complex killing mechanism activated by B. thuringiensis and its pore-forming toxins. B. thuringiensis toxins produce midgut lesions that are the entry sites for B. thuringiensis spores and enteric microbes into the hemocoel, where they are hindered by immune barriers. The rationale behind our experimental approach was to finely modulate these barriers and assess the resulting effect on septicemia and mortality levels associated with a constant level of damage to the midgut epithelium. This approach allowed us to study, in a realistically physiological context characterized by an unaltered midgut microbiota, the role of the enteric flora in the induction of lethal septicemia following B. thuringiensis toxin-induced damage. This is a controversial hypothesis that has been widely debated over the last decade (22, 23, 27, 28, 40).
The adoption of invasive experimental protocols (i.e., the use of antibiotics associated with the artificial delivery of selected bacteria), which will have a profound effect on the midgut microbiota and possibly on administered B. thuringiensis (25, 32), has raised serious concerns regarding the biological relevance of the reported results and the value of their interpretation. The unnatural conditions associated with elimination of the bacterial flora will have a major impact on a wealth of functions, including immune defense barriers, pathogen vectoring capacity, tissue regeneration, and, more generally, physiological homeostasis, with a significant influence on nutrition and detoxification pathways (33, 41–45). The experimental approach adopted here did not alter the midgut environment in any detectable way, but simply acted on the defense response by selectively decreasing the transcript level of a gene expressed in the hemocytes, which controls cellular immunity. This allowed a down-regulation of immunocompetence without using immunosuppressive molecules (26, 34), which may have unintended effects on a range of physiological functions and on their cross-modulation with defense reactions (46–48).
The results obtained with this targeted approach clearly indicate that the experimentally immunosuppressed larvae are far more sensitive to B. thuringiensis infection, as well as to its toxin Cry1Ca. Therefore, the limited capacity of bacterial clearance because of 102 Sl gene silencing enhanced the septicemia induced by B. thuringiensis toxin ingestion. The changes occurring in the microbiota of the hemolymph strongly corroborate this hypothesis. Indeed, the experimental data demonstrate that midgut bacteria overcome the intestinal barrier, ulcerated by B. thuringiensis pore-forming toxins, and reach the hemocoel, where bacterial proliferation is toxin dose-dependent. These effects were clearly enhanced by RNAi-mediated immunosuppression, which caused the same rates of mortality and septicemia at nearly fivefold lower doses of B. thuringiensis toxin compared with immunocompetent controls. The microbiota in the hemocoel of experimental larvae exposed to an LC50 dose of B. thuringiensis toxin was similar to that of the midgut, characterized by higher relative abundance of Serratia. This is in accordance with previous reports on the pathogenic role of enterobacteria when they enter the hemocoel after B. thuringiensis treatment, switching from a gut symbiont to a systemic pathogen (22, 28). Overall, the in vivo observations reported here support the hypothesis that the septicemia in host larvae exposed to B. thuringiensis toxin plays a key role in the induction of mortality. Moreover, the similar increase in susceptibility induced by host immunosuppression in both the presence and absence of B. thuringiensis spores indicates that the gut microbiota has an important role in the complex virulence strategy adopted by this entomopathogen.
The present study provides a functional framework that may account in part for the reported discrepancies of the impact exerted by B. thuringiensis toxins on different insect populations (1, 2). These differences could be explained by the heterogeneous microbiota composition and differing degrees of host immunocompetence, which are influenced by various biotic and abiotic factors. Indeed, the capacity of insects to mount a costly defense reaction is affected by multiple factors, including nutritional status, developmental phase, co-occurrence of multiple pathogens and parasites, as well as by any other competing metabolic response to environmental stress agents (49, 50).
Our comparative analysis of the microbiota occurring in the midgut and in the hemolymph revealed distinct differences. The hemocoel microbiota of control larvae was characterized by a completely unique profile, with higher relative abundances of taxonomic groups barely represented in the midgut. This clearly indicates that without the tissue lesions induced by B. thuringiensis toxins, these are separate compartments in which different microbial communities with completely different functional roles are established and maintained. The unexpectedly low presence of bacteria in the hemocoel of larvae not exposed to B. thuringiensis or its toxin was not due to experimental manipulation to deliver dsRNA, as supported by data obtained from unmanipulated healthy larvae, showing nearly identical bacterial loads and microbiota composition of experimental controls (SI Appendix, Fig. S6 and Table S5). Moreover, the limited presence of bacterial species not associated with septicemia in the hemolymph of healthy larvae, experimental controls, and larvae exposed to a low toxin dose could be due either to a selective passage through the gut wall or to a differential clearance following hemocoel entrance. A selective passage through the midgut epithelium could be a possible way to allow a controlled transfer of bacteria to reproductive tissues and eggs. In this way, the egg and newly hatched larvae could receive an early supply of the important microbial complement, which indeed seems to be already present in the eggs of Lepidoptera (51). This reasonable speculation is indirectly supported by a recent study reporting the transit of ingested bacteria into the hemolymph and their accumulation in the ovary (52). Given the important role of the gut microbiota in the overall physiological balance (33, 42, 53), the occurrence of a transmission mechanism would not be surprising.
In conclusion, the present study provides a clear picture of the complex interplay among B. thuringiensis toxins, the host caterpillar, and its midgut microbiota. By providing insight into these interactions, we have shown clear-cut evidence regarding the important role of host septicemia in the multifaceted host killing mechanism mediated by the B. thuringiensis toxin. However, the importance of host septicemia does not exclude the possible involvement of other negative effects related to the disruption of gut structural and functional integrity that may synergistically contribute to host death by negatively affecting the metabolism and, indirectly, the immunocompetence. Moreover, these results have important implications from an applied perspective, setting the stage for the development of selective RNAi-based strategies of insect pest control aimed at enhancing the impact of natural antagonists by reducing the immunocompetence of the host.
Methods
Gene Silencing.
Silencing of the 102 Sl gene by RNAi and the assessment of its transcription level by qRT-PCR were performed as described previously (35).
Toxicity Bioassays.
At 6 h after the last dsRNA administration, newly molted fifth instar larvae were isolated and used for LC50 calculation of both B. thuringiensis and Cry1Ca, according to the protocol described in SI Appendix, Methods.
Nodulation and Phagocytosis Assays.
For the nodulation assay, at 12 h after the last dsRNA administration (SI Appendix, Fig. S7), S. littoralis larvae, surface-sterilized with 70% ethanol and chilled on ice, received an intrahemocoelic injection of 2 × 106 E. coli cells suspended in 5 µL of PBS (137 mM NaCl, 2.7 mM KCl, and 10 mM phosphate buffer, pH 7.4). Injections were performed through the neck membrane using a Hamilton 1701 RN SYR (10 µL, 26s gauge, 55 mm long, point style 3). A thoracic leg was cut 18 h after injection, and the exuding hemolymph was collected and immediately diluted into an equal volume of ice-cold Mead anticoagulant buffer (98 mM NaOH, 145 mM NaCl, 17 mM EDTA, and 41 mM citric acid, pH 4.5). The hemocyte nodules occurring in the hemolymph samples were counted (n = 28) under a microscope at 400× magnification (Axioskop; Carl Zeiss Microscopy), using a Bürker chamber. Nodulation assays on larvae exposed to Cry1Ca treatment were performed similarly at the same experimental time points (n = 5–10 on each sampling point) used for qRT-PCR quantification of bacterial loads (SI Appendix, Fig. S7). When an intense immune response gave rise to large aggregates of nodules difficult to count separately, the number of distinct nodules observed was arbitrarily doubled, because the percentage of nonwhite pixels measured (ZEN software; Carl Zeiss Microscopy) on the large aggregates was on average twice that measured on a bright-microscopy field containing discrete nodules and free hemocytes.
Phagocytic activity by S. littoralis hemocytes was assessed in vivo at 12 h after the last dsRNA administration, by injecting 10 µL of a PBS suspension of 2 × 107 fluorescein-conjugated E. coli cells (K-12 strain BioParticles, fluorescein conjugate; Life Technologies), reconstituted according to the manufacturer’s instructions. After 15 min, the hemolymph was collected from a cut thoracic leg into ice-cold PBS (2:1) and then loaded into a Bürker chamber. The total number of hemocytes and of hemocytes showing fluorescence were determined using a fluorescence microscope (Axioskop 20; Carl Zeiss Microscopy). Dead hemocytes (2.02 ± 0.05%; n = 10) were counted at the end of incubation in a Bürker chamber by a trypan blue exclusion assay on a sample aliquot mixed (2:1, vol:vol) with 0.4% (wt:vol in water) trypan blue (Sigma-Aldrich).
Humoral Antimicrobial Response.
The humoral immune response as affected by gene silencing was assessed by measuring the level of transcription of genes encoding antimicrobial peptides and lysozymes in response to bacterial injections (SI Appendix, Fig. S7). In brief, 6 h after the last dsRNA administration, S. littoralis larvae, surface-sterilized with 70% ethanol and chilled on ice, received an intrahemocoelic injection of 2 × 107 E. coli or 3 × 108 Staphylococcus aureus cells, suspended in 5 µL of PBS. Injections were performed through the neck membrane with a Hamilton 1701 RN SYR (10 µL, gauge 26s, length 55 mm, needle 3). At the time of injection (T0) and at 18 h after injection (T1), larvae (n = 8 for each experimental sample) were dissected and hemocytes, midgut, and fat body were collected and processed for total RNA extraction, as described previously (35). The relative expression of attacin 1, gloverin, and lysozyme 1a were assessed by q-RT-PCR, as described in SI Appendix, Methods.
Cry1Ca Toxin Treatment and Collection of Samples for Microbiota Analysis.
At 6 h after the last dsRNA administration, newly molted fifth instar larvae of S. littoralis, processed to silence the 102 Sl gene (35), were exposed for 3 d to 0.2 and 1.2 µg/cm2 of Cry1Ca, whereas control larvae, treated with GFP dsRNA, received higher toxin doses of 1.2 and 6.6 µg/cm2. Both experimental groups included internal controls maintained on a toxin-free diet. The experimental doses used were determined by taking into account the different level of susceptibility associated with gene silencing to compare treatments having similar impacts in terms of mortality, such as LC50, and doses approximately fivefold lower than that value. On day 7, larvae were transferred to an untreated diet, and 24 h later, the midgut content and hemolymph (∼30 µL per larva) were collected separately under a horizontal laminar flow hood (SI Appendix, Fig. S7). Experimental samples were obtained by pooling 10 larvae. The experiment was repeated three times.
To collect the midgut content, the larva was cut longitudinally, and the explanted midgut was opened lengthwise with microscissors. The material released from the lumen was immediately homogenized with a pestle in a tube containing PBS (50 µL per midgut content), vigorously vortexed, and centrifuged at 2,900 × g for 10 min at 4 °C. The recovered supernatant was split into two aliquots and used for studying the microbiota (i.e., bacterial counts and 16S rRNA gene-targeted metagenomics).
A thoracic leg was cut, and the exuding hemolymph was collected and centrifuged at 300 × g for 10 min at 4 °C to remove the hemocytes. The plasma thus obtained was split into two aliquots for subsequent analyses of the microbiota as above.
Analysis of the Microbiota.
Total RNA was immediately extracted from samples using a PowerMicrobiome RNA Isolation Kit (MO BIO Laboratories), according to the manufacturer’s recommendations. The concentration of extracted RNA was assessed by measuring the absorbance at 260 nm with a Varioskan Flash Multimode Reader (Thermo Fisher Scientific), and its purity was evaluated by assessing the 260 nm/280 nm absorbance ratio. Total RNA preparations were then treated with TURBO DNase I (Life Technologies), according to the manufacturer’s instructions. For each sample, 200 ng of bacterial RNA was reverse-transcribed into cDNA with random primers using RETROscript (Life Technologies), according to the manufacturer’s instructions.
The structure of the microbiota in the midgut and hemolymph samples was assessed by pyrosequencing of the amplified V1–V3 region of the 16S rRNA gene as described previously (54). The bioinformatics analysis was conducted as described in SI Appendix, Methods. The 16S rRNA gene sequences are available at the National Center for Biotechnology Information’s Sequence Read Archive (accession no. SRP064613).
Changes over time in the relative bacterial load in the midgut (n = 5 for each sampling point) and hemolymph (n = 10–12 for each sampling point) samples, collected as described above, were determined by qRT-PCR (SI Appendix, Methods) to assess the impact of different doses of Bt toxin and 102 Sl silencing (SI Appendix, Fig. S7). In addition, viable counts of the midgut and hemolymph microbiota were performed by plating 1:10 dilutions of the samples onto LB agar, followed by incubation at 25 °C for 48 h.
Light Microscopy and Transmission Electron Microscopy.
The methodology is described in detail in SI Appendix, Methods.
Statistical Analysis.
Data were analyzed using GraphPad Prism version 6.0b and SPSS version 21 (IBM). Experiments with two groups were analyzed using the unpaired Student's t test, whereas experiments with more than two groups were analyzed using one-way ANOVA and Tukey’s multiple-comparison test. Two-way ANOVA was carried out on AMP and lysozyme transcriptional analysis, as affected by dsRNA treatment and bacterial injection, and three-way ANOVA was performed to compare the relative quantification data of bacteria and nodule formation as affected by dsRNA treatment, time, and Cry1Ca toxin exposure. Levene’s test was used to test the homogeneity of variance. When necessary, transformation of data was carried out to meet the assumptions of normality and homoscedasticity. When significant effects were observed (P < 0.05), the Bonferroni post hoc test was used to compare mean values.
Supplementary Material
Acknowledgments
We thank Angharad Gatehouse (University of Newcastle) and Salvador Herrero (University of Valencia) for their comments on the manuscript; and Gennaro Di Prisco (University of Napoli Federico II) for his help and suggestions. This work was supported by the Ministero dell’Istruzione, dell’Università e della Ricerca, Futuro in Ricerca 2013 (RBFR13PMT1), the POR Campania Fondo Europeo Sviluppo Regionale 2007–2013 Progetto BIP, and the Cluster Agroalimentare Nazionale SAFE&SMART Project (CTN01_00230_248064).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (accession no. SRP064613).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521741113/-/DCSupplemental.
References
- 1.van Frankenhuyzen K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol. 2009;101(1):1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.van Frankenhuyzen K. Cross-order and cross-phylum activity of Bacillus thuringiensis pesticidal proteins. J Invertebr Pathol. 2013;114(1):76–85. doi: 10.1016/j.jip.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Palma L, Muñoz D, Berry C, Murillo J, Caballero P. Bacillus thuringiensis toxins: An overview of their biocidal activity. Toxins (Basel) 2014;6(12):3296–3325. doi: 10.3390/toxins6123296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz de Escudero I, et al. A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. J Invertebr Pathol. 2014;117:51–55. doi: 10.1016/j.jip.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Soberón M, et al. Pore formation by Cry toxins. Adv Exp Med Biol. 2010;677:127–142. doi: 10.1007/978-1-4419-6327-7_11. [DOI] [PubMed] [Google Scholar]
- 6.Pardo-López L, Soberón M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: Mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev. 2013;37(1):3–22. doi: 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- 7.Roh JY, Choi JY, Li MS, Jin BR, Je YH. Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J Microbiol Biotechnol. 2007;17(4):547–559. [PubMed] [Google Scholar]
- 8.Bravo A, Likitvivatanavong S, Gill SS, Soberón M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol. 2011;41(7):423–431. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S, Chandra A, Pandey KC. Bacillus thuringiensis (Bt) transgenic crop: An environment-friendly insect-pest management strategy. J Environ Biol. 2008;29(5):641–653. [PubMed] [Google Scholar]
- 10.Bravo A, Soberón M. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 2008;26(10):573–579. doi: 10.1016/j.tibtech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Storer NP, Thompson GD, Head GP. Application of pyramided traits against Lepidoptera in insect resistance management for Bt crops. GM Crops Food. 2012;3(3):154–162. doi: 10.4161/gmcr.20945. [DOI] [PubMed] [Google Scholar]
- 12.Carrière Y, Crickmore N, Tabashnik BE. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015;33(2):161–168. doi: 10.1038/nbt.3099. [DOI] [PubMed] [Google Scholar]
- 13.Ferré J, Van Rie J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu Rev Entomol. 2002;47:501–533. doi: 10.1146/annurev.ento.47.091201.145234. [DOI] [PubMed] [Google Scholar]
- 14.Ferré J, Van Rie J, MacIntosh SC. Insecticidal genetically modified crops and insect resistance management (IRM) In: Romeis J, Shelton AM, Kennedy GG, editors. Integration of Insect-Resistant Genetically Modified Crops Within IPM Programs. Springer; Dordrecht, The Netherlands: 2008. pp. 41–85. [Google Scholar]
- 15.Tabashnik BE, Brévault T, Carrière Y. Insect resistance to Bt crops: Lessons from the first billion acres. Nat Biotechnol. 2013;31(6):510–521. doi: 10.1038/nbt.2597. [DOI] [PubMed] [Google Scholar]
- 16.Jakka S, Ferré J, Jurat-Fuentes JL. Cry toxin binding site models and their use in strategies to delay resistance evolution. In: Soberón M, Gao Y, Bravo A, editors. Bt Resistance: Characterization and Strategies for GM Crops Producing Bacillus thuringiensis Toxins. Centre for Agriculture and Biosciences International; Oxfordshire, UK: 2015. pp. 138–149. [Google Scholar]
- 17.González-Cabrera J, et al. Toxicity and mode of action of Bacillus thuringiensis Cry proteins in the Mediterranean corn borer, Sesamia nonagrioides (Lefebvre) Appl Environ Microbiol. 2006;72(4):2594–2600. doi: 10.1128/AEM.72.4.2594-2600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernández-Rodríguez CS, Hernández-Martínez P, Van Rie J, Escriche B, Ferré J. Shared midgut binding sites for Cry1A.105, Cry1Aa, Cry1Ab, Cry1Ac, and Cry1Fa proteins from Bacillus thuringiensis in two important corn pests, Ostrinia nubilalis and Spodoptera frugiperda. PLoS One. 2013;8(7):e68164. doi: 10.1371/journal.pone.0068164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández-Martínez P, et al. Shared binding sites for the Bacillus thuringiensis proteins Cry3Bb, Cry3Ca, and Cry7Aa in the African sweet potato pest Cylas puncticollis (Brentidae) Appl Environ Microbiol. 2014;80(24):7545–7550. doi: 10.1128/AEM.02514-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen-LeRoux C, Gaudriault S, Ramarao N, Lereclus D, Givaudan A. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr Opin Microbiol. 2012;15(3):220–231. doi: 10.1016/j.mib.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Castagnola A, Stock SP. Common virulence factors and tissue targets of entomopathogenic bacteria for biological control of lepidopteran pests. Insects. 2014;5(1):139–166. doi: 10.3390/insects5010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick NA, Raffa KF, Handelsman J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc Natl Acad Sci USA. 2006;103(41):15196–15199. doi: 10.1073/pnas.0604865103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broderick NA, et al. Contributions of gut bacteria to Bacillus thuringiensis-induced mortality vary across a range of Lepidoptera. BMC Biol. 2009;7:11. doi: 10.1186/1741-7007-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston PR, Crickmore N. Gut bacteria are not required for the insecticidal activity of Bacillus thuringiensis toward the tobacco hornworm, Manduca sexta. Appl Environ Microbiol. 2009;75(15):5094–5099. doi: 10.1128/AEM.00966-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raymond B, et al. A mid-gut microbiota is not required for the pathogenicity of Bacillus thuringiensis to diamondback moth larvae. Environ Microbiol. 2009;11(10):2556–2563. doi: 10.1111/j.1462-2920.2009.01980.x. [DOI] [PubMed] [Google Scholar]
- 26.Broderick NA, Raffa KF, Handelsman J. Chemical modulators of the innate immune response alter gypsy moth larval susceptibility to Bacillus thuringiensis. BMC Microbiol. 2010;10:129. doi: 10.1186/1471-2180-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Frankenhuyzen K, Liu Y, Tonon A. Interactions between Bacillus thuringiensis subsp. kurstaki HD-1 and midgut bacteria in larvae of gypsy moth and spruce budworm. J Invertebr Pathol. 2010;103(2):124–131. doi: 10.1016/j.jip.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Mason KL, et al. From commensal to pathogen: Translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta. MBio. 2011;2(3):e00065–e11. doi: 10.1128/mBio.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patil CD, Borase HP, Salunke BK, Patil SV. Alteration in Bacillus thuringiensis toxicity by curing gut flora: Novel approach for mosquito resistance management. Parasitol Res. 2013;112(9):3283–3288. doi: 10.1007/s00436-013-3507-z. [DOI] [PubMed] [Google Scholar]
- 30.Paramasiva I, Sharma HC, Krishnayya PV. Antibiotics influence the toxicity of the delta endotoxins of Bacillus thuringiensis towards the cotton bollworm, Helicoverpa armigera. BMC Microbiol. 2014;14:200. doi: 10.1186/1471-2180-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visweshwar R, Sharma HC, Akbar SMD, Sreeramulu K. Elimination of gut microbes with antibiotics confers resistance to Bacillus thuringiensis toxin proteins in Helicoverpa armigera (Hubner) Appl Biochem Biotechnol. 2015;177(8):1621–1637. doi: 10.1007/s12010-015-1841-6. [DOI] [PubMed] [Google Scholar]
- 32.Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. Bacillus thuringiensis: An impotent pathogen? Trends Microbiol. 2010;18(5):189–194. doi: 10.1016/j.tim.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Buchon N, et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Reports. 2013;3(5):1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Richards EH, Dani MP. A recombinant immunosuppressive protein from Pimpla hypochondriaca (rVPr1) increases the susceptibility of Lacanobia oleracea and Mamestra brassicae larvae to Bacillus thuringiensis. J Invertebr Pathol. 2010;104(1):51–57. doi: 10.1016/j.jip.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Di Lelio I, et al. Functional analysis of an immune gene of Spodoptera littoralis by RNAi. J Insect Physiol. 2014;64:90–97. doi: 10.1016/j.jinsphys.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32(10):1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 37.Falabella P, et al. Functional amyloids in insect immune response. Insect Biochem Mol Biol. 2012;42(3):203–211. doi: 10.1016/j.ibmb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Yi H-Y, Chowdhury M, Huang Y-D, Yu X-Q. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98(13):5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 40.Grizanova EV, Dubovskiy IM, Whitten MMA, Glupov VV. Contributions of cellular and humoral immunity of Galleria mellonella larvae in defence against oral infection by Bacillus thuringiensis. J Invertebr Pathol. 2014;119:40–46. doi: 10.1016/j.jip.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109(27):11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 43.Soumana IH, et al. The bacterial flora of tsetse fly midgut and its effect on trypanosome transmission. J Invertebr Pathol. 2013;112(Suppl):S89–S93. doi: 10.1016/j.jip.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 44.Carissimo G, et al. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc Natl Acad Sci USA. 2015;112(2):E176–E185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ceja-Navarro JA, et al. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat Commun. 2015;6:7618. doi: 10.1038/ncomms8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolowczuk I, et al. Feeding our immune system: Impact on metabolism. Clin Dev Immunol. 2008;2008:639803. doi: 10.1155/2008/639803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci USA. 2009;106(49):20853–20858. doi: 10.1073/pnas.0906749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tracey KJ. Reflex control of immunity. Nat Rev Immunol. 2009;9(6):418–428. doi: 10.1038/nri2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ponton F, et al. Integrating nutrition and immunology: A new frontier. J Insect Physiol. 2013;59(2):130–137. doi: 10.1016/j.jinsphys.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 50.Nazzi F, Pennacchio F. Disentangling multiple interactions in the hive ecosystem. Trends Parasitol. 2014;30(12):556–561. doi: 10.1016/j.pt.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Hernández-Martínez P, et al. Increase in midgut microbiota load induces an apparent immune priming and increases tolerance to Bacillus thuringiensis. Environ Microbiol. 2010;12(10):2730–2737. doi: 10.1111/j.1462-2920.2010.02241.x. [DOI] [PubMed] [Google Scholar]
- 52.Freitak D, et al. The maternal transfer of bacteria can mediate trans-generational immune priming in insects. Virulence. 2014;5(4):547–554. doi: 10.4161/viru.28367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douglas AE. The molecular basis of bacterial-insect symbiosis. J Mol Biol. 2014;426(23):3830–3837. doi: 10.1016/j.jmb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ercolini D, De Filippis F, La Storia A, Iacono M. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl Environ Microbiol. 2012;78(22):8142–8145. doi: 10.1128/AEM.02218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.