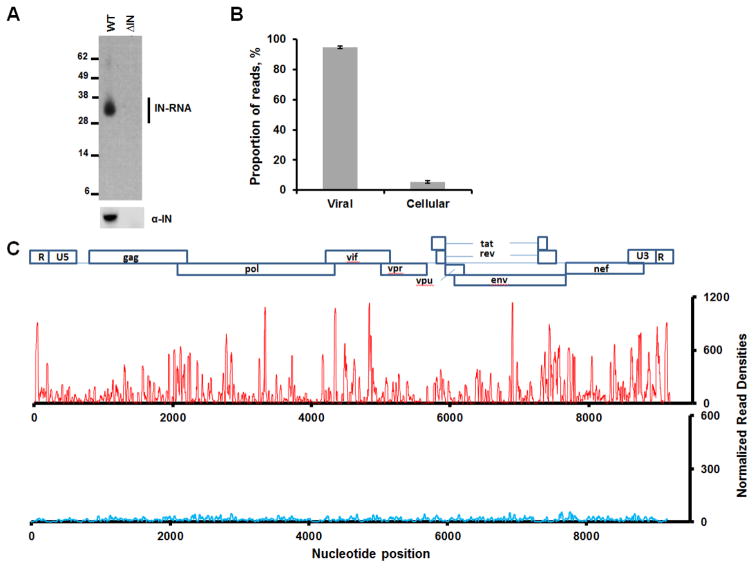
Figure 1. CLIP-seq experiments showing HIV-1 IN binding to viral RNA in virions.
(A) A representative autoradiogram of IN-RNA crosslinked adducts from HIV-1NL4-3 and HIV-1NL4-3 ΔIN virions (upper panel). Corresponding western blot analysis of IN in immunoprecipitated fractions (lower panel). (B) Percentage of reads that map to cellular and viral RNA obtained from 2 independent CLIP-Seq analysis of IN-RNA adducts from WT virions. Error bars indicate standard errors. (C) CLIP-seq results showing read densities (frequency distribution of nucleotide occurrence) mapped to the viral genome obtained from HIV-1NL4-3 (red) and HIV-1NL4-3 ΔIN (blue) progeny virions. The read densities have been normalized with respect to total reads for WT HIV-1NL4-3. A collinear schematic diagram of the HIV-1 genome is shown above.