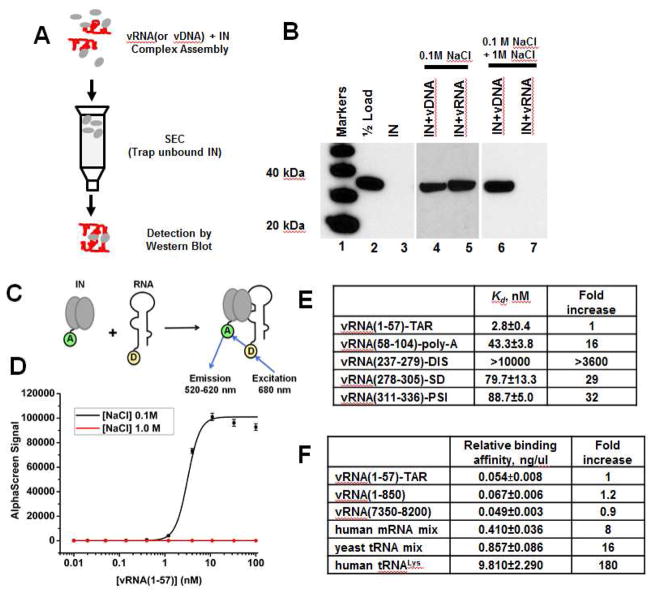
Figure 2. HIV-1 IN binding to synthetic viral RNA segments in vitro.
(A) Schematic for the SEC approach to test IN interactions with nucleic acids. (B) SDS-PAGE and immunoblotting with anti-IN antibodies of SEC fractions. Lane 1: molecular weight markers; Lane 2: 1/2 input IN; Lane 3: unliganded IN subjected to SEC; Lanes 4 and 5: IN+vDNA or IN+vRNA(1-850) complexes were assembled in the buffer containing 0.1 M NaCl and subjected to SEC in the same buffer. Lanes 6 and 7: IN+vDNA or IN+vRNA(1-850) complexes were formed in the buffer containing 0.1 M NaCl. Then NaCl concentration was increased to 1M followed by SEC. (C) AlphaScreen assay design to monitor direct binding between 6xHis-tagged WT IN and biotinylated RNA oligonucleotides (only IN dimer shown for simplicity). “A” and “D” indicate anti-His Acceptor and streptavidin coated Donor beads bound to 6xHis-tagged IN or biotin respectively. (D) Representative AlphaScreen binding curves for IN binding to vRNA(1-57)-TAR in the presence of 0.1 M or 1 M NaCl. The average values of three independent experiments and corresponding standard deviations are shown. (E and F) Summary of AlphaScreen-based assays for IN binding affinities for various RNA oligonucleotides with standard errors of three independent experiments shown.