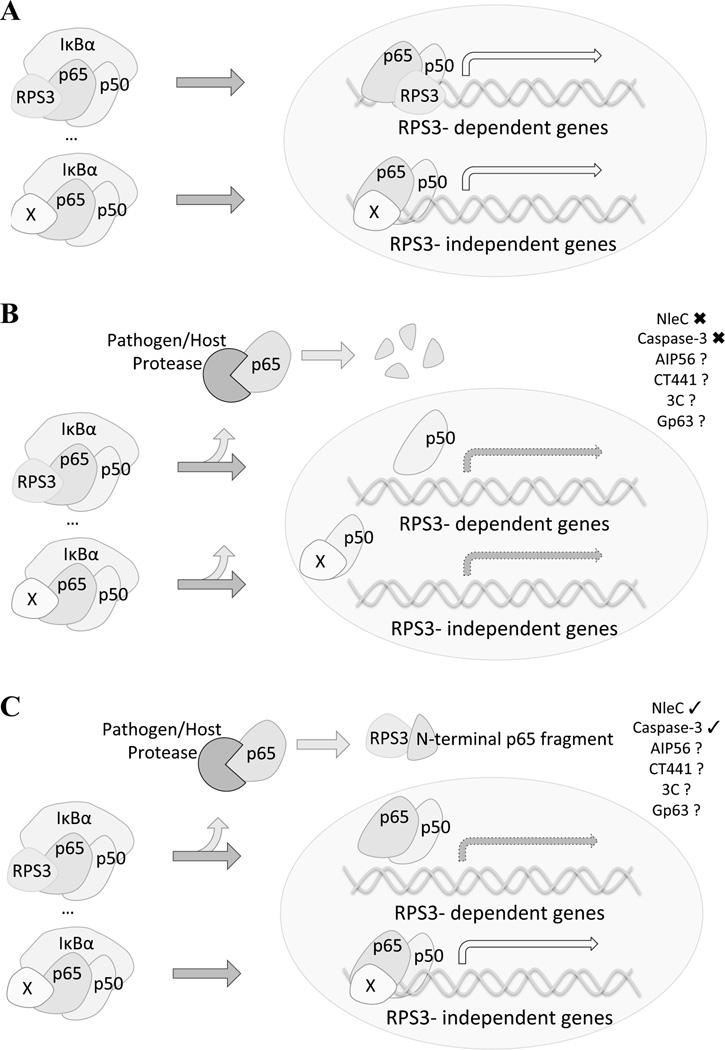
Figure 1.
Comparison of global and selective inhibition interpretation models of pathogen-encoded protease mediated NF-κB interference. Under infection conditions, NF-κB activation results in p65 nuclear translocation and the transactivation of a wide array of target genes, a subset of which require the aid of known specifier proteins, such as RPS3 and Sam68. This leads to a coordinated cellular responses and clearance of infection (A). Pathogen-encoded proteases can have devastating, widespread effects (global, B) or they can inhibit specific aspects of a pathway. Complete cleavage of p65 interrupts the signal transduction cascade, preventing all downstream outcomes in a global fashion. However, partial proteolytic processing of p65 can result in selective inhibition (C), as there are still some full length and functioning p65 molecules within the cell. In the experimentally verified (checked) examples of NleC-mediated cleavage of p65, during A/E pathogen infections, an N-terminal fragment is produced that sequesters RPS3, preventing the activation of that arm of the NF-κB signaling pathway. This inhibition thus results in the amplification of the effect of cleaving only a percentage of the p65 cellular pool. It is likely, yet remains to be verified experimentally (question marked), that a similar mechanism is employed by other pathogens. EPEC/EHEC/C. rodentium- NleC; Phdp- AIP56; C. trachomatis- CT441; Poliovirus- 3C; Leishmania spp.- Gp63; Host- Caspase-3.