Figure 1.
In Vivo Identification of RBDs by RBDmap
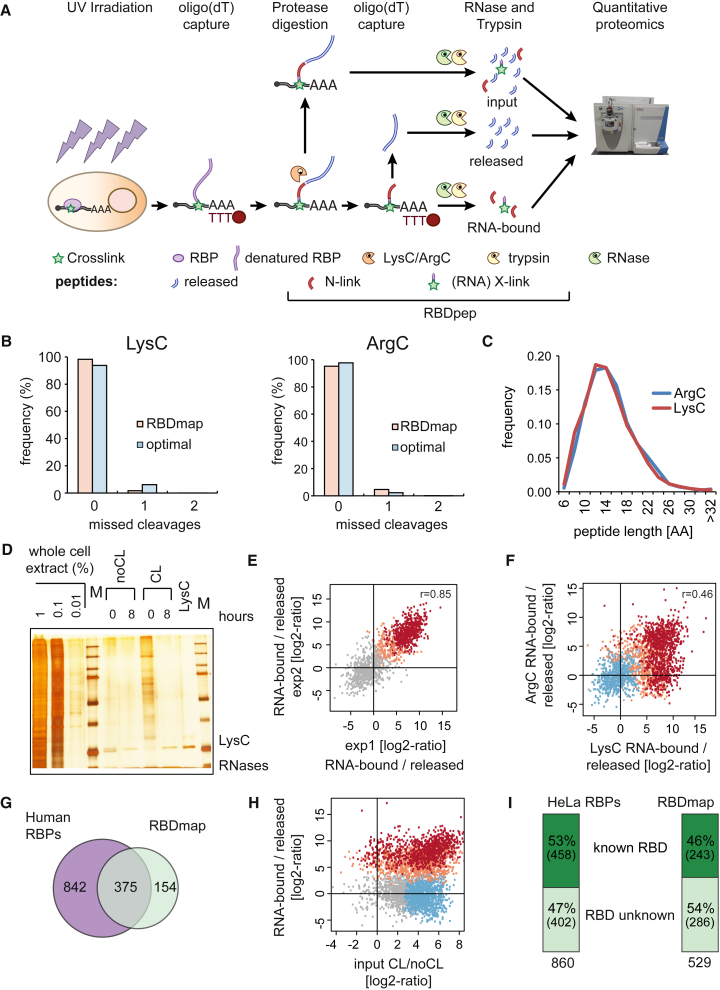
(A) Schematic representation of the RBDmap workflow.
(B) LysC- and ArgC-mediated proteolysis was monitored without trypsin treatment. The protease digestion under RBDmap conditions or in buffers typically used in MS studies (optimal) were compared to in silico digestions defining 0% miscleavage. The missed cleavages were calculated and plotted.
(C) Distribution of MS-identified LysC/ArgC fragments based on their number of amino acids.
(D) Silver staining shows the protein pattern of purified RBPs prior to and after LysC treatment (crosslinking: CL).
(E) Scatter plot comparing the peptide intensity ratios between RNA-bound and released fractions. The peptides enriched in the RNA-bound fraction at 1% (RBDpep) and 10% FDR (candidate RBDpep) are shown in red and salmon, respectively (Pearson correlation coefficient: r).
(F) Peptide intensity ratios between LysC and ArgC experiments computed from three biological replicates. The dots represent released peptides (blue), RBDpeps (red), candidate RBDpeps (salmon), and background peptides (gray).
(G) Venn diagram comparing the proteins within the RBDmap data set and the HeLa, HEK293, and Huh-7 RNA interactomes.
(H) Comparison of the peptide intensity ratios from three biological replicates between UV-irradiated and non-irradiated inputs (x axis) and between RNA-bound and released fractions (y axis) (color code as above).
(I) Number of proteins harboring recognizable or unknown RBDs in the HeLa mRNA interactome (left) and in RBDmap dataset (right).