Figure 4.
Globular RBDs Discovered by RBDmap
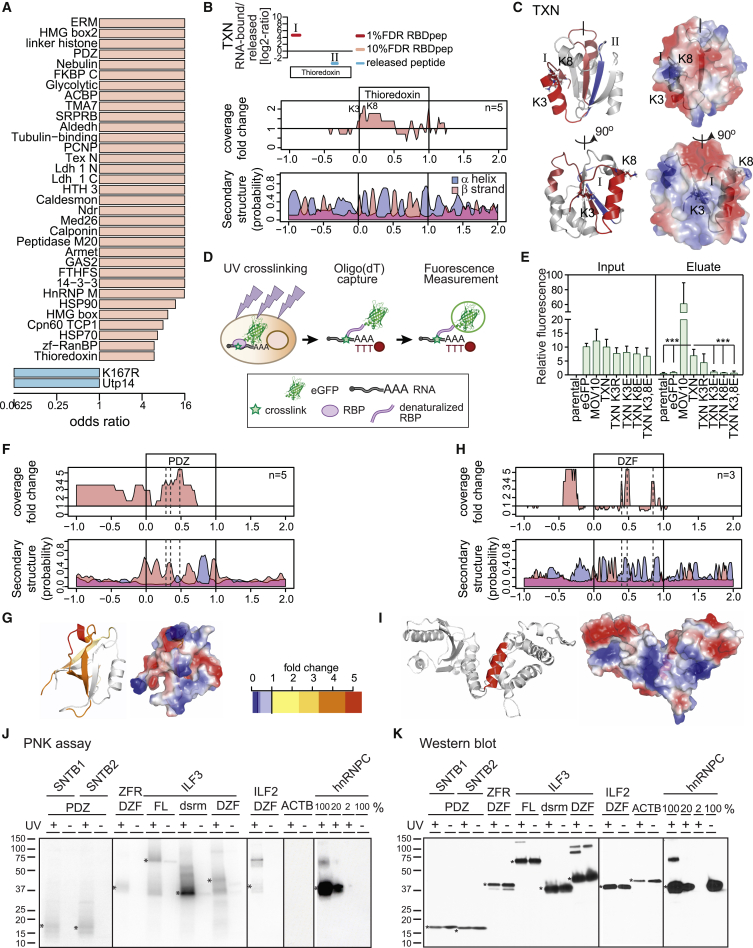
(A) Odds ratios for the most highly enriched RBDs.
(B) RBDpep and released peptides mapping to TXN as in Figure 2D (top). The ratio of the X-link over released peptide coverage at each position of the TXN fold as in Figure 3C is shown (middle). The secondary structure prediction for each position of the TXN fold and flanking regions is shown (bottom).
(C) Crystal structure of human TXN (PDB 3M9J), K3 and K8 are highlighted, and the identified RBDpep is shown in red.
(D) Schematic representation of the protocol for measurement of RNA-binding using eGFP fusion proteins.
(E) Relative total (input) or RNA-bound (eluate) green fluorescence signal from cells expressing different eGFP fusion proteins (∗∗∗p < 0.01, t test, and n = 9).
(F) As in (B), but for PDZ domain.
(G) Ratio of X-link over released peptides plotted as a heatmap in a PDZ homology model.
(H) As in (B), but for DZF domain.
(I) As in (G), but using a DZF homology model.
(J) Autoradiography of FLAG-HA tagged proteins after PNK assay.
(K) Western blotting using an antibody against the HA tag. The polypeptides of the expected molecular masses are indicated by asterisks.