FIGURE 5.
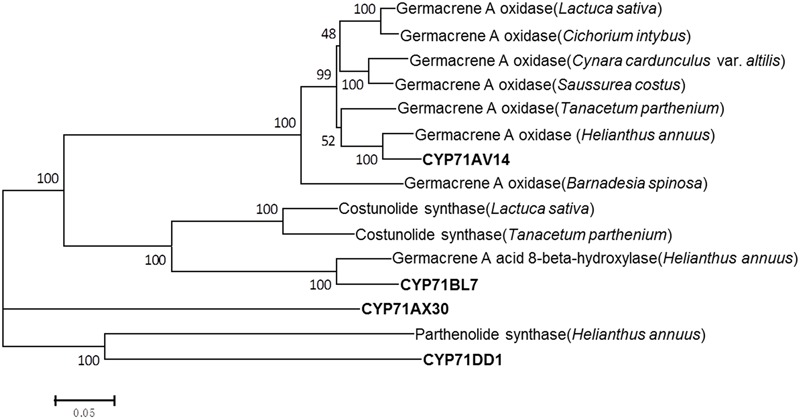
Phylogenetic analysis of the putative CYP71 candidates of this study with the previously published known GAA or GAA-derived oxidizing enzymes. Amino acid sequences were aligned using Clustal X software. The tree was inferred by the neighbor-joining method using MEGA 6.0 software. The scale bar represents 0.2 amino acid substitutions per site. Numbers indicate the bootstrap values of 1000 replicates. Germacrene A oxidase (Helianthus annuus), ADF43082.1; germacrene A oxidase (Tanacetum parthenium), AHN62855.1; germacrene A oxidase (Saussurea costus), ADF43081.1; germacrene A oxidase (Cynara cardunculus var. altilis), AIA09036.1; germacrene A oxidase (Lactuca sativa), ADF32078.1; germacrene A oxidase (Cichorium intybus), ADF43080.1; germacrene A oxidase (Barnadesia spinosa), ADF43083.1; costunolide synthase (Lactuca sativa), AEI59780.1; costunolide synthase (Tanacetum parthenium), AGO03790.1; germacrene A acid 8-beta-hydroxylase (Helianthus annuus), F8S1H3.1; parthenolide synthase (Helianthus annuus), AHM24033.1.
