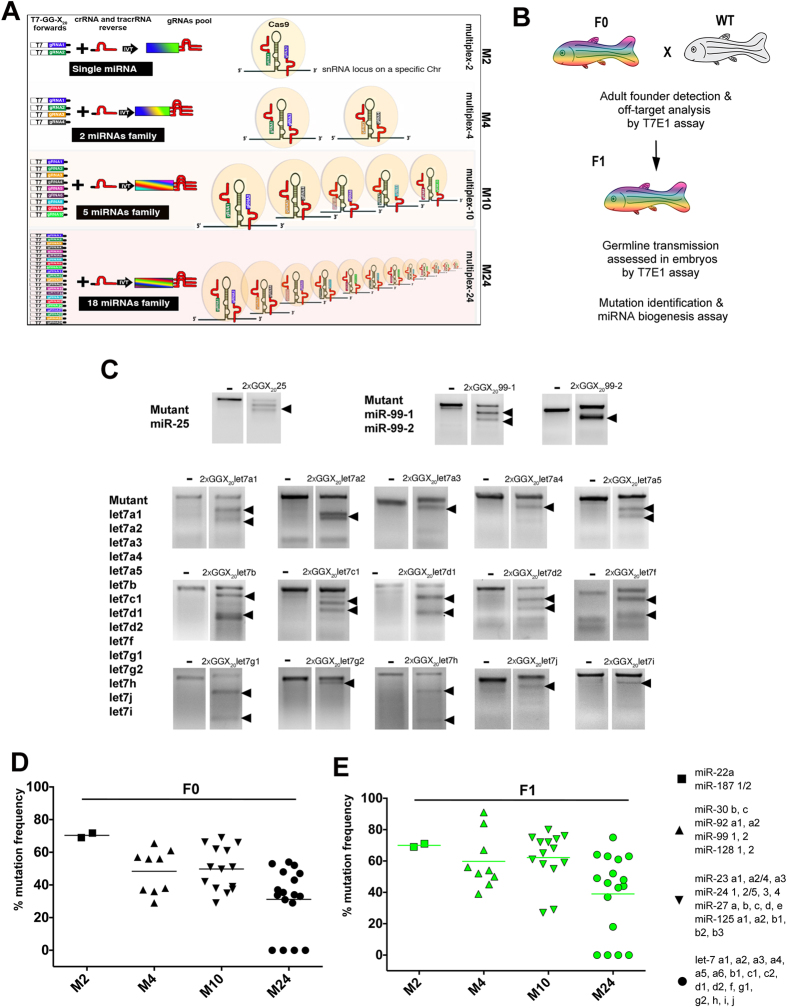
Figure 1. Multiplex CRISPR/Cas9 strategy to target miRNA families.
(A) Diagram indicating the design of multiplexed sgRNAs against the miRNA family members. Pairs of forward gRNA primers were customized for each target miRNA and consisted of a 5′-T7 RNA sequence followed by GG-X20 (X20 represents the 20 nt gene specific sgRNA and the black line represents a generic 15 nt sequence for annealing with the universal reverse primer). Double-stranded DNAs containing sgRNA sequences were assembled using annealing and PCR amplification with a universal reverse primer containing crRNA and tracrRNA sequences. Pooled sgRNAs were synthetized in a single in vitro transcription reaction (IVT). Multiplexing (M) was accomplished to target a miRNA family and/or duplicates, consisting of 1, 2, 5, and 18 miRNAs. For the let-7 family, 24-multiplexed sgRNAs were sufficient to target 36 loci since multiple let-7 members share an identical pre-miRNA genome sequence. (B) Schematic representing the strategy to generate miRNA family mutants. (C) F0 fish injected with the respective multiplexed sgRNAs and Cas9 mRNA were genotyped using gene specific primers that amplified a 200-400 nt genomic region for one, two, and up to 18 miRNA loci. CRISPR/Cas9 mutagenesis was determined by the T7EI assay. Cleaved PCR fragments revealed the presence of indels as indicated with arrowheads. Uninjected wild type fish (minus sign) were used as negative controls. Mutation frequencies were determined by T7EI assay in genotyped F0 adult zebrafish (D) and F1 progeny (E) and quantified as previously described19,22. Each data point represents the average mutation frequency of miRNA target loci analyzed by T7E1 assay in an individual F0 adult zebrafish (D) or F1 embryo (E). At least five individual adult F0 fish (D) and twenty-five F1 embryos (E) were screened for each multiplexed sgRNA pool (Mn).