Fig. 8.
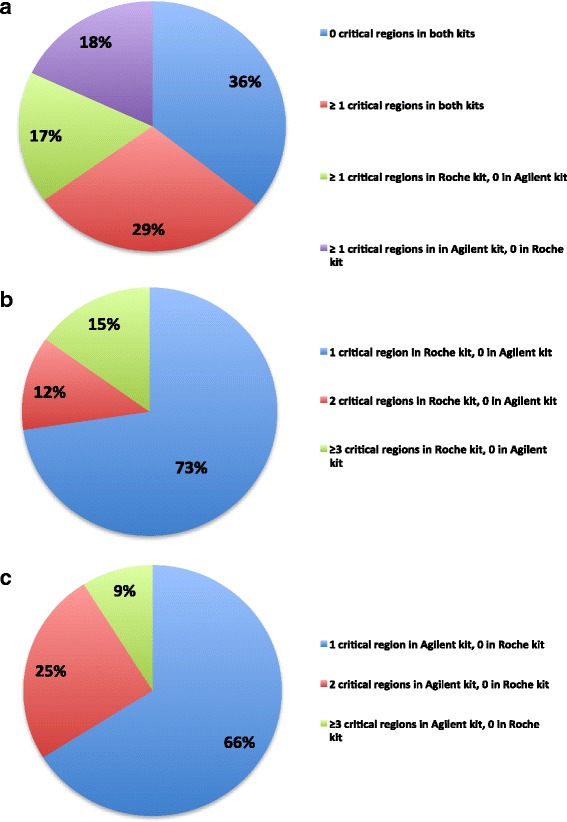
Coverage distribution across all the coding exons of 623 cancer-related genes in both WES platforms. Distribution summary of 623 cancer-related genes according to their coverage performance achieved in the two tested WES systems (a). Specifically, 36 % of the genes (red) were completely well covered by both Agilent and Roche kits; 29 % (blue) had at least one ‘critical’ region in both kits; 18 % were completely well covered by Roche NimbleGen kit, but had one or more ‘critical’ region in Agilent SureSelect kit; finally, 17 % of the genes were completely well covered by Agilent SureSelect kit, but had one or more problematic region in Roche NimbleGen kit. Distribution summary of cancer-related genes having one (73 %), two (12 %) or more (15 %) critical regions in NimbleGen Roche kit, but completely well-covered in Agilent SureSelect kit (b). Distribution summary of cancer-related genes having one (66 %), two (25 %) or more (9 %) critical regions in Agilent SureSelect kit, but completely well-covered in Roche NimbleGen kit (c)
