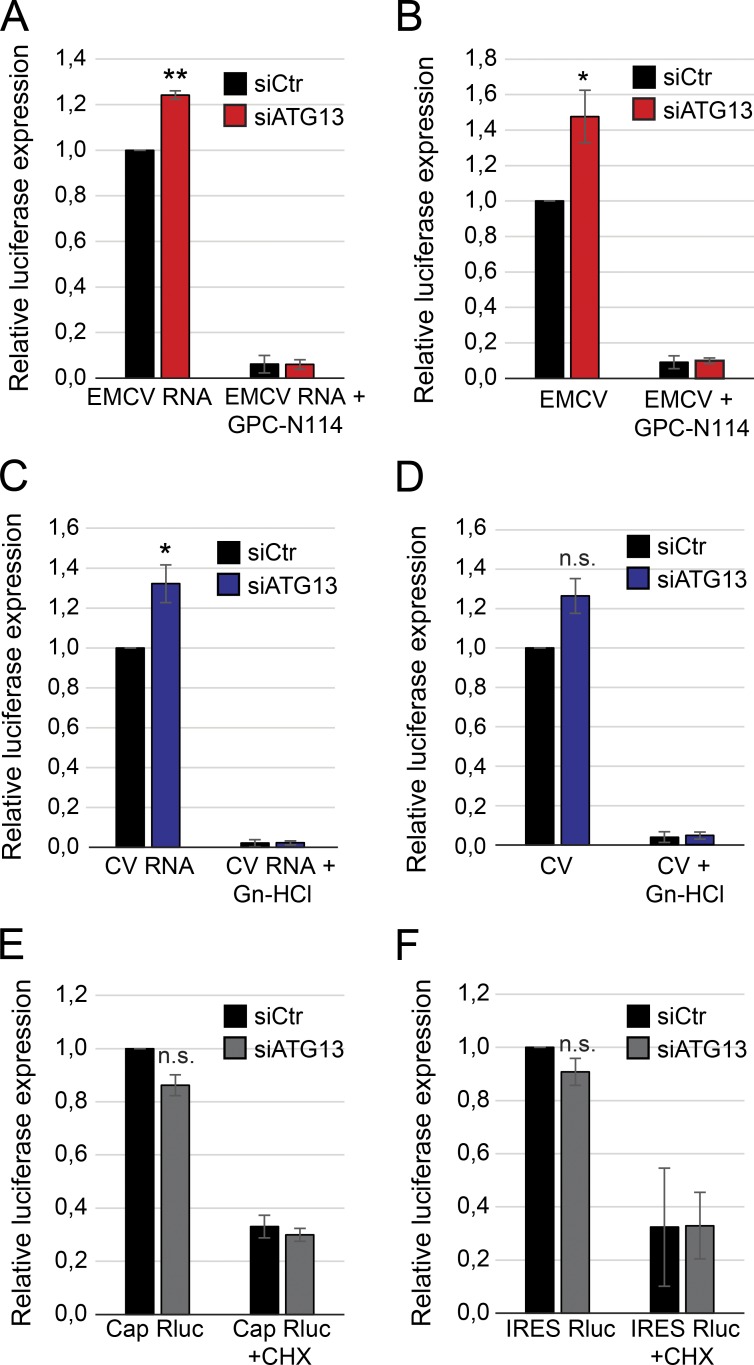
Figure 7.
ATG13 depletion enhances EMCV and CV replication, but not virus cell entry. (A) U2OS cells depleted or not of ATG13 using siRNA for 48 h were transfected with Renilla luciferase EMCV RNA for 7 h. GPC-N114 was added or not 1 h after virus inoculation before measuring luciferase expression (n = 3). (B) Cells prepared as in A were infected with luciferase-expressing EMCV for 7 h. GPC-N114 was added or not 1 h after virus inoculation before assessing luciferase expression (n = 5). (C) Cells prepared as in A were transfected with Renilla luciferase CV RNA for 7 h. Guanidine-HCL (Gn-HCl) was added or not 1 h posttransfection before measuring luciferase expression (n = 4). (D) Cells prepared as in A were infected with luciferase expressing CV for 7 h. Gn-HCl was added or not 1 h after virus inoculation before assessing luciferase expression (n = 4). (E) Cells were transfected with the cap RNA Renilla-luciferase (Rluc) transcript for 7 h and, when indicated, cycloheximide (CHX) was added 1 h after transfection before determining luciferase expression (n = 3). (F) Cells were transfected with the IRES-luciferase RNA transcript for 7 h and when indicated, CHX was added 1 h after transfection before measuring luciferase expression. (n = 3). All data are presented relative to the control (folds). Error bars represent SEM. *, P < 0.05; **, P < 0.01; n.s., not significant. siCtr, scramble siRNA.