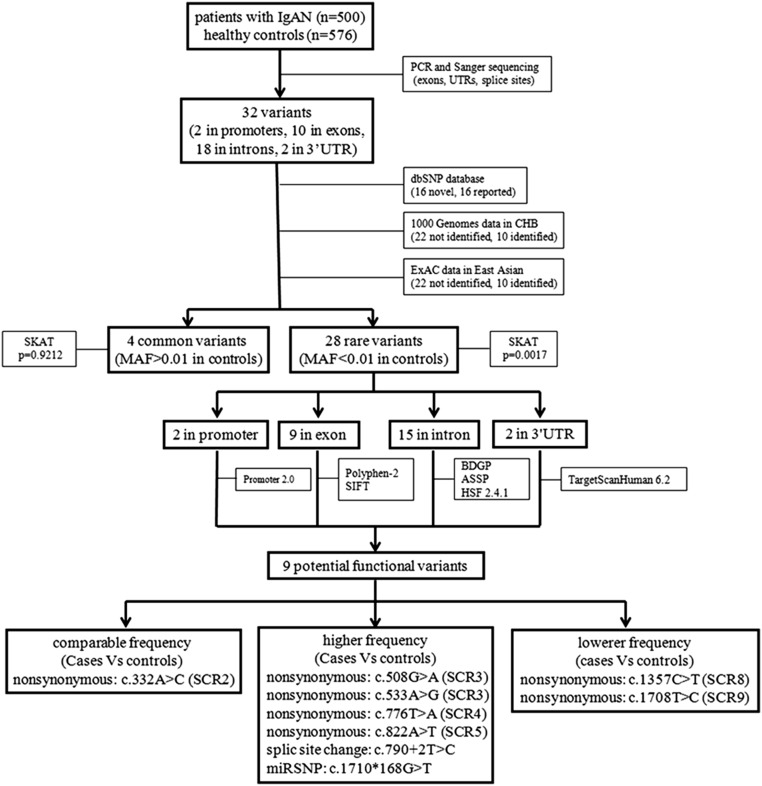
Figure 1.
Flowchart of the genetic analysis of CFHR5 variants in IgAN. After screening for variants in CFHR5 in 500 patients with IgAN and 576 healthy controls, 32 variants were identified. After checking against the dbSNP database, 1000 Genome data, and ExAC data to see whether these variants were reported before, 32 variants in CFHR5 were divided into two groups, common variants and rare variants, on the basis of the MAF in controls (cutoff =0.01). Then, variant distribution between patients with IgAN and controls was compared in each group using the SKAT. After the identification of rare variants in CFHR5 associated with IgAN, the functional meaning of these rare variants was predicted by in silico programs. At last, nine potential functional variants were identified, which were located in different SCRs of the CFHR5 protein. CHB, Chinese Han Beijing; dbSNP, single nucleotide polymorphism database; ExAC, exome aggregation consortium; HSF, human splicing finder; SIFT, sorting intolerant from tolerant.