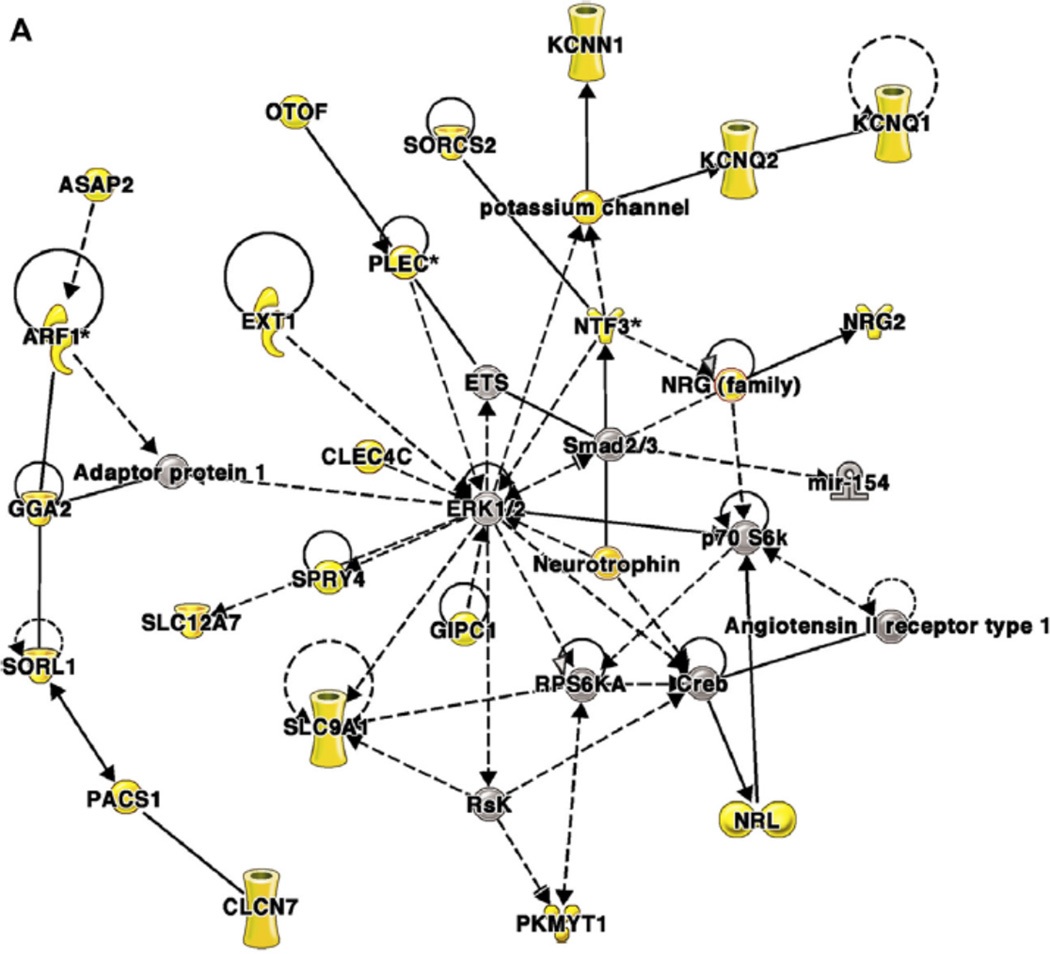
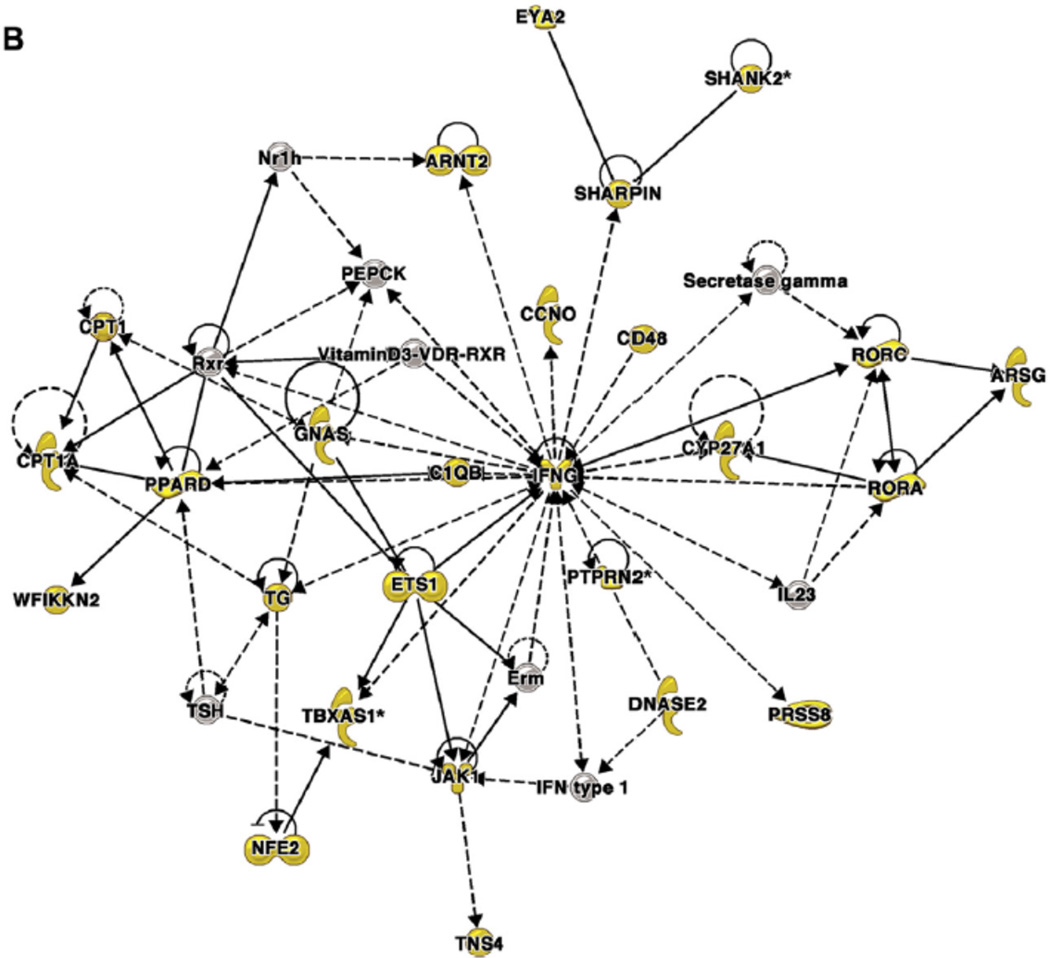
Figure 3. Coexpression network analysis of genes near differentially methylated CpG dinucleotides in airway epithelial cells from asthmatic and nonasthmatic subjects84.
Networks were generated by using Ingenuity Pathway Analysis (IPA), as previously described.84 Genes near differentially methylated CpGs are shown as yellow symbols; other genes are included to connect the network of differentially methylated CpGs. Molecular shapes (from IPA) were as follows: oval, transcriptional regulator; diamond, enzyme; dashed line rectangle, channel; upward triangle, phosphatase; downward triangle, kinase; trapezoid, transporter; circle, other. A, A network from module 1 that was associated with clinical markers of asthma severity. This network is centered on extracellular signal-regulated kinase 1/2, which was not itself near a differentially methylated CpG. B, A network from module 2 that was associated with eosinophilia. This network is centered on IFN-g, which was near a differentially methylated CpG. Reprinted with permission of the American Thoracic Society from Nicodemus-Johnson et al.84 Copyright 2016 American Thoracic Society.