Abstract
Two retrotransposons from the superfamilies Copia and Gypsy named as Copia-LTR_SS and Gypsy-LTR_SS, respectively, were identified in the genomic bank of Sclerotinia sclerotiorum. These transposable elements (TEs) contained direct and preserved long terminal repeats (LTR). Domains related to codified regions for gag protein, integrase, reverse transcriptase and RNAse H were identified in Copia-LTR_SS, whereas in Gypsy-LTR_SS only domains for gag, reverse transcriptase and RNAse H were found. The abundance of identified LTR-Solo suggested possible genetic recombination events in the S. sclerotiorum genome. Furthermore, alignment of the sequences for LTR elements from each superfamily suggested the presence of a RIP (repeat-induced point mutation) silencing mechanism that may directly affect the evolution of this species.
Keywords: phytopathogens, retrotransposons, transposable elements
Transposable elements (TEs) are ubiquitous DNA sequences in the genome that have the ability to move from one place to another (Kidwell, 2005). TEs form two classes based on the transposition mechanisms involved: class I includes the TEs usually referred to as retrotransposons and class II contains the "DNA transposons" per se. All class I TEs are transposed by intermediate RNA which is transcribed from a copy of the genome and the cDNA is obtained from a reverse transcriptase codified by the element itself. Every complete transposon cycle produces a new copy and, consequently, retrotransposons are frequently the main contributors to a repetitive fraction of the genome. Retrotransposons can be classified in five groups based on their mechanism of transposition and on the organization and phylogeny of the reverse transcriptase: LTR (long terminal repeat), DIRS-like (Dictyostelium intermediate repeat sequence), Penelope-like, LINEs (long interspersed nuclear elements) and SINEs (short interspersed nuclear elements) (Wicker et al., 2007).
LTR retrotransposons are usually found in fungi, especially in the superfamilies Gypsy and Copia. The LTR gag and pol regions are structural compounds of the Copia and Gypsy retrotransposons. The LTRs flank the 5' and 3' extremities that are identical with active retrotransposons. The gag region encodes structural proteins similar to those of the viral capsid. The pol region encodes a polyprotein that is processed to yield the proteins involved in the transposition. These proteins include a protease involved in protein maturation and cleavage, a reverse transcriptase that reverse-transcribes the RNA into cDNA, an integrase that allows transposon insertion into the genome, and an RNAse H that degrades the RNA regions during cDNA synthesis. In addition, the PPT (polypurine tract) and PBS (primer binding site) regions facilitate transposon transcription in the genus (Havecker et al., 2004; Manetti et al., 2007). The Gypsy and Copia retrotransposons differ from each other in the arrangement of the sequence that encodes the reverse transcriptase and integrase (Wicker et al., 2007).
TEs activities in the genome may affect gene structure and its regulation (Shapirova, 2010; Huan-Van et al., 2011). In addition, TEs provide important sites for ectopic recombination in the genome (Dean et al., 2005; Ohm et al., 2012). In this regard, the genomes of organisms have different strategies to avoid possible damage caused by TEs present in the genome, including a silencing mechanism known as RIP (repeat-induced point mutation) that was originally discovered in Neurospora crassa (Selker, 1990, 2002). RIP occurs during the sexual cycle, between fertilization and karyogamy, and induces GC-to-AT mutations in duplicated DNA sequences longer than 400 pb and with an identity of > 80% (Galagan and Selker, 2004).
In most fungal species, TEs generally represent 2-20% of the genome, although in some cases they can account for 85% of the genome (Parlange et al., 2011). Transposons are important elements for evolution of the genome in phytopathogenic fungi because of their linked gain or loss of virulence (Khang et al., 2008; Chuma et al., 2011). Many effector genes in plant pathogens occur in genomic regions that are rich in TEs. These ETs may alter the gene structure or expression and stimulate the emergence of new pathogenic races (Bakkeren and Valent, 2014). In addition, the presence of cognate-TEs in conserved domains of genes can lead to their integration into regulatory reticulations via microRNA (Li et al., 2011).
Sclerotinia sclerotiorum, the causal agent of white mold, has a worldwide distribution with a range of hosts that consists of at least 408 species and 278 plant genera. Analyses of the genetic diversity of S. sclerotiorum TEs have suggested recent genomic remodeling involving TE expansion (Amselem et al., 2011; Santana et al., 2014a). The S. sclerotiorum genome is estimated to contain 38 Mb, 7% of which consists of TEs, with the frequency of LTR-retrotransposons being ~2-2.5% (Amselem et al., 2011). In this work, we investigated the possible evolutionary impacts of TEs in the S. sclerotiorum genome.
The genomic sequences of S. sclerotiorum class I transposable elements were obtained by searching the fungal genome database (http://www.ncbi.nlm.nih.gov/assembly/GCF_000146945.1/) and using the LTR-Finder software. Subsequently, the remaining copies of the elements were obtained by using the Basic Local Alignment Search Tool (BLAST) for each previously identified element against the S. sclerotiorum genome. The main domains related to TE-encoded regions were tagged with the BLASTX tool (http://www.ncbi.nlm.nih.gov). TEs were classified based on their structural features and by phylogenetic sequence analysis that encoded the reverse transcriptase protein. The neighbor-joining method with a bootstrap value of 5,000 replications was used for the phylogenetic analysis and included the reverse transcriptase protein sequence from different TE groups: Maggy from Magnaporthe grisea (AAA33420), Real from Alternaria alternata (BAA89272), Ty3 from Saccharomyces cerevisiae (M23367), copia from Drosophila simulans (D10880), Ty1 from S. cerevisiae (Z48149), jockey from Drosophila melanogaster (M22874), Penelope from Drosophila virilis (AAL14979) and DIRS from Lytechinus variegatus (BK001257). The sequence alignment and phylogenetic analysis were done using MEGA4 software (Tamura et al., 2007).
Evidence for a RIP silencing mechanism was obtained from an analysis of 157 sequences from Copia-LTR_SS retrotransposons and 12 sequences from Gypsy-LTR_SS retrotransposons. The sequences were aligned using MEGA4 software (Tamura et al., 2007). The dinucleotide frequency analysis and estimation of the RIP indices were determined using RipCal software (Hane and Oliver, 2008). The indices or ratios used to prove RIP were TpA/ApT and (CpA+TpG)/(ApC+GpT). The data obtained for S. sclerotiorum were compared to the transposase sequences of Colletrotrichum cereale (Crouch et al., 2008), the PeTra element of Penicillium chrysogenum (Braumann et al., 2008), element OPUIO3-1414 of Ophiostoma novo-ulmi (Bouvet et al., 2008), element Fot 1 of Fusarium oxysporum (Daboussi et al., 1992) and element Punt of N. crassa (Magolin et al., 1998) by using the same indices.
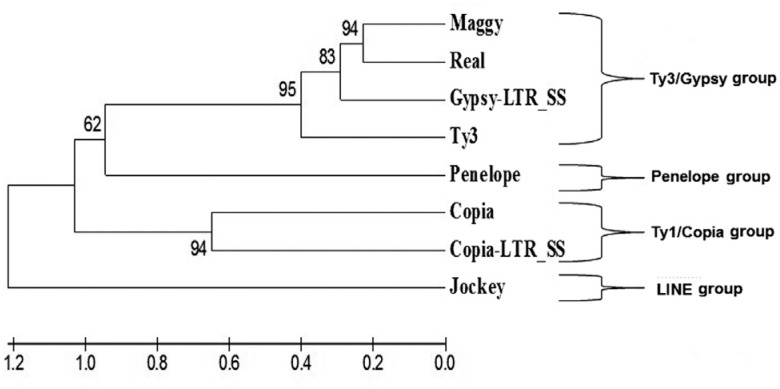
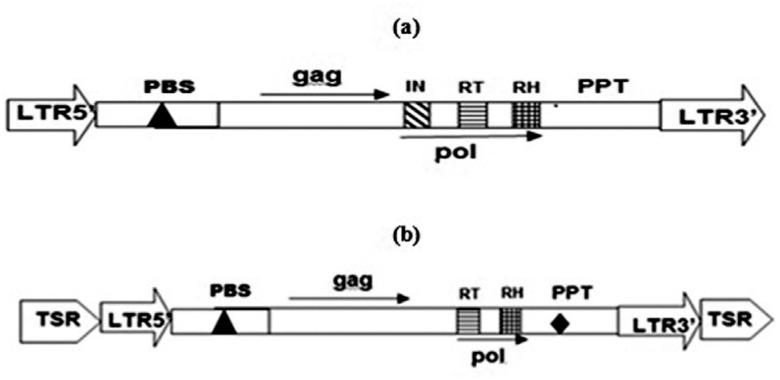
Examination of the S. sclerotiorum genome revealed two retrotransposons possibly involved in the restructuring of the fungal genome. The structural and phylogenetic analyses (Figure 1) of these two elements allowed their classification as part of the superfamilies Copia and Gypsy; the elements were referred to as Copia-LTR_SS (supercontig 8: 23.5003-24.0346) and Gypsy-LTR_SS (supercontig 36: 1.308-7.775) (Figure S1 (55.4KB, pdf) , Supplementary material). A total of seven complete elements were identified, six of which belonged to the Copia element and one to the Gypsy element. The Copia-LTR_SS element (5,344 bp) ` long terminal repetitions (LTRs) that were directly conserved (269 bp) and conserved domains that encoded gag and pol region proteins such as integrase, reverse transcriptase and RNAse H (Figure 2A).
Figure 1. A dendrogram showing the grouping of the Copia-LTR_SS and Gypsy-LTR_SS elements. The analysis was done using the neighbor-joining method based on 5,000 bootstrap replicates. The numbers above and below each node indicate the percentage of times in which each branch appeared in a bootstrap analysis with 5,000 replicates. X-axis numbers refer to genetic divergence.
Figure 2. Structural organization of the retrotransposons Copia-LTR_SS (A) and Gypsy-LTR_SS (B) identified in the genome of S. sclerotiorum. The pol region of Copia-LTR_SS contained domains for integrase (IN), reverse transcriptase (RT) and RNAse H (RH), whereas Gypsy-LTR_SS had only RT and RH domains. The two elements had PBS (primer binding site) and PPT (polypurine tract) regions. Large arrows represent the LTRs.
The type of protein and its position in the open-reading frame (ORF) were typical of elements from the superfamily Copia. The conserved LTRs (435 bp) and the conserved domain for the Gag protein, the reverse transcriptase and the RNAse H were also found in the Gypsy-LTR_SS element (6,468 bp). Nonetheless, the domain containing the integrase and protease were not labeled in this element (Figure 2B). Mutations in this element are a possible explanation for the absence of the integrase and protease domains and will result in an inactive element. In addition, most of the retrotransposon sequences found in the S. sclerotiorum genome are degenerate (Amselem et al., 2011). The conserved Gypsy-LTR_SS contained LTR 5' and 3' flanking insertion signs known as TSRs (target site repeats) that consisted of five base-pairs (GAAAT). These TSRs are duplicated TE target sequences that arise at the moment of insertion. In both elements, purine-rich regions known as PPT and PBS were identified. These regions are important for the reverse transposons of TE. An analysis of approximately 5,000 bp in the upstream and downstream sequences of the complete TEs demonstrated that the elements occurred in regions rich in repetitive sequences and were neighbors to genes related to mRNA splicing, apoptosis and heterokaryon incompatibility.
In all, 141 and 359 solo-LTR sequences were identified for the Copia-LTR_SS and Gypsy-LTR_SS retrotransposons, respectively. The presence of non-autonomous elements and solo-LTRs in S. sclerotiorum highlighted the possible occurrence of ectopic recombination in this fungal genome. The reason for this is that these sequences generally result in recombination between TE sequences and those of the same family. Additional evidence for recombination in the S. sclerotiorum genome involving TEs was the fact that the different TSRs flanked copies of the six identified Copia-LTR_SS elements. The presence of different insertion areas in the extremities of a single TE may reflect the recombination of similar retrotransposons containing different TSRs. Ectopic recombination through transposons has been reported as an important genome reconstruction event in many fungi, such as Magnaporthe grisea (Dean et al., 2005), Coprinopsis cinerea (Stajich et al., 2010), Verticilium dahliae (Amyotte et al., 2012), Mycosphaerella fijiensis (Santana et al., 2012) and Cochliobolus heterostrophus (Santana et al., 2014b), among others.
TRIM (terminal-repeat retrotransposon in miniature) elements were also tagged: three sequences originated from the Copia-LTR_SS element and 21 from the Gypsy-LTR_SS element. These transposons result from autonomous LTR retroelements. However, the DNA sequences related to pol or gag region proteins are absent, making these elements defective (non-autonomous). However, these elements can move through the genome using the enzymatic machinery of similar elements (Wicker et al., 2007).
The results of RIP analysis of the LTR sequences of Copia-LTR_SS and Gypsy-LTR_SS TEs were compared to those reported in the literature and their corresponding RIP mechanism (Table 1). The ratios TpA/ApT and (CpA + TpG)/(ApC+GpT) obtained for S. sclerotiorum were the same as those already reported in the literature. This finding suggested that the existing CpA-dinucleotides in the LTRs of suitable elements in S. sclerotiorum are the target of mutations generated by a process similar to the RIP mechanism. Evidence of RIP in S. sclerotiorum has also been provided by Amselem et al. (2015). In contrast, Santana et al. (2014a) found no RIP silencing mechanism in Tc1-Mariner elements (class II transposons) of the S. sclerotiorum genome. Together, these findings indicate variation in the occurrence of RIP silencing mechanisms among TEs in S. sclerotiorum. Similar behavior has been reported for the genomes of Aspergillus niger (Braumann et al., 2008) and C. heterostrophus (Santana et al., 2014b).
Table 1. TpA/ApT and CpA+TpG/ApC+GpT ratios for transposons and retrotransposons.
| Sequences analyzed | (TpA/ApT) | (CpA+TpG)/(ApC+GpT) | Reference |
|---|---|---|---|
| Retrotransposon Copia-LTR_SS | 1.35 | 0.27 | This study |
| Retrotransposon Gypsy-LTR_SS | 1.0 | 0.95 | This study |
| Transposase (Colletrotrichum cereale) | 2.00 | 0.44 | Crouch et al. (2008) |
| PetTra (Penicillium chrysogenum) | 1.22 | 0.58 | Braumann et al. (2008) |
| OPHIO3-1414 (Ophiostoma novo-ulmi) | 1.51 | 0.60 | Bouvet et al. (2008) |
| Fot1 (Fusarium oxysporum) | 1.12 | 0.75 | Daboussi et al. (1992) |
| Punt (Neurospora crassa) | 1.32 | 0.56 | Magolin et al. (1998) |
Standard index values for RIP are (TpA/ApT) > 0.89 and (CpA+TpG)/(ApC+GpT) < 1.03, (www.sourceforge.net/protects/ripcal).
The presence of a RIP silencing mechanism in the genome of phytopathogenic fungi may have a significant impact on the evolution of these organisms. For instance, in C. heterostrophus the mutation site is located in the transposons and in regions near the TEs (Ohm et al., 2012). This mechanism may accelerate the rate of evolution in this genus, depending on the number of effector genes that are located close to TE-rich regions (Grandaubert et al., 2014). Gene duplication is important for the evolution of a species and the presence of a RIP mechanism may have a significant impact on the evolution of several fungi. For example, the presence of a RIP mechanism is associated with the absence or paucity of duplicated genes in the N. crassa genome. In addition to creating one or more copies of a functional gene, a RIP silencing mechanism may also generate new alleles. Indeed, this mechanism is regarded as essential for the emergence of genes with new functions (Galagan and Selker, 2004).
In conclusion, TEs may play an important role in organizing the S. sclerotiorum genome and can potentially increase the adaptation of this species to different environments and hosts. Such adaptation makes control of the disease more difficult. Furthermore, the abundance of copia solo-LTR and TRIMs identified in S. sclerotiorum should facilitate the use of these sequences as molecular markers in future investigations of genetic variability in this fungus.
Acknowledgments
Financial support by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Instituto de Biotecnologia Aplicada à Agropecuária (BIOAGRO) of the Universidade Federal de Viçosa is acknowledged.
Supplementary Material
The following online material is available for this article:
This material is available as part of the online article from http://www.scielo.br/gmb
Footnotes
Associate Editor: Louis Bernard Klaczko
References
- Amselem J, Cuomo J, van Kan JAL, Viaud M, Benito EP, Coulox A, Coutinho PM, Vries RP, Dyer PS, Fillinger S, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 2011;7:1–27. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J, Lebrun M-H, Quesneville H. Whole gnome comparative analysis of transposable elements provides new insight into mechanisms of their inactivation in fungal genomes. BMC Genomics. 2015;16: doi: 10.1186/s12864-015-1347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyotte SG, Tan X, Pennerman K, Jimenez-Gasco MM, Klosterman SJ, Ma LJ, Dobinson KF, Veronese P. Transposable elements in phytopathogenic Verticillium spp.: Insights into genome evolution and inter- and intra-specific diversification. BMC Genomics. 2012;13: doi: 10.1186/1471-2164-13-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren G, Valent B. Do pathogen effectors play peek-a-boo? Front Plant Sci. 2014;5:1–2. doi: 10.3389/fpls.2014.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet FB, Jacob B, Plourd KV, Bernier L. Stress-induced mobility of OPHO1 and OPHO2, DNA transposons of the Dutch elm disease fungi. Fungal Genet Biol. 2008;45:565–578. doi: 10.1016/j.fgb.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Braumann I, Berg M, Klempken F. Repeat induced point mutation in two asexual fungi, Aspergillus niger and Penicillium chrysogenum . Curr Genet. 2008;53:287–297. doi: 10.1007/s00294-008-0185-y. [DOI] [PubMed] [Google Scholar]
- Chuma I, Isobe C, Hotta Y, Ibaragi K, Futamata N, Kusaba M, Yoshida K, Terauchi R, Fujita Y, Nakayashiki H, et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011;7: doi: 10.1371/journal.ppat.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch JA, Glasheen BM, Giunta MA, Clarke BB, Hillman BI. The evolution of transposon repeat-induced point mutation in the genome of Colletrotrichum cereale: Reconciling sex, recombination and homoplasy in an "asexual" pathogen. Fungal Genet Biol. 2008;45:190–206. doi: 10.1016/j.fgb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Daboussi MJ, Langin T, Brygoo Y. Fot1, a new family of fungal transposable elements. Mol Gen Genet. 1992;232:2–16. doi: 10.1007/BF00299131. [DOI] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kullarni R, Xu JR, Pan H, et al. The genome sequence of the rice blast fungus Magnaporthe grisea . Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU. RIP: The evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Grandaubert J, Balesdent M-H, Rouxel T. Evolutionary and adaptive role of transposable elements in fungal genomes. Adv Bot Res. 2014;70:79–107. [Google Scholar]
- Hane J, Oliver RP. RIPCAL: A tool for alignment-based analyses of repeat-induced point mutations in fungal genomics sequences. BMC Bioinform. 2008;9:478–478. doi: 10.1186/1471-2105-9-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol. 2004;5:225–225. doi: 10.1186/gb-2004-5-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan-Van A, Rouzic AL, Boutin TS, Filée J, Capy P. The struggle for life of the genome's selfish architects. Biol Direct. 2011;6:19–19. doi: 10.1186/1745-6150-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang CH, Park S-Y, Lee Y-H, Valent B, Kang S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol Plant Microbe Int. 2008;21:658–670. doi: 10.1094/MPMI-21-5-0658. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements. In: Gregory TR, editor. The Evolution of the Genome. Elsevier Academic Press; San Diego: 2005. pp. 105–221. [Google Scholar]
- Li Y, Li C, Xia J, Jin Y. Domestication of transposable elements into microRNA genes in plants. PLoS One. 2011;6: doi: 10.1371/journal.pone.0019212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magolin BS, Garrett-Engele PW, Stevens JN, Fritz DY, Garrett-Engele C, Metzengerg RL, Selker EU. A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics. 1998;149:1787–1791. doi: 10.1093/genetics/149.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti ME, Rossi M, Costa APP, Clausen AM, Van Sluys M. Radiation of the Tnt I retrotransposon superfamily in three Solanaceae genera. BMC Evol Biol. 2007;7: doi: 10.1186/1471-2148-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, et al. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen dothideomycetes fungi. PLoS Pathog. 2012;8: doi: 10.1371/journal.ppat.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlange F, Oberhaensli S, Breen J, Platzer M, Taudien S, Simková H, Wicker T, Dolezel J, Keller B. A major invasion of transposable elements accounts for the large size of the Blumeria graminis f.sp. tritici genome. Funct Integr Genomics. 2011;11:671–677. doi: 10.1007/s10142-011-0240-5. [DOI] [PubMed] [Google Scholar]
- Santana MF, Silva JCF, Batista AD, Ribeiro LE, Silva GF, Araújo EF, Queiroz MV. Abundance, distribution and potential impact of transposable elements in the genome of Mycosphaerella fijiensis . BMC Genomics. 2012;13: doi: 10.1186/1471-2164-13-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana MF, Silva JCF, Mizubuti ESG, Araújo EF, Queiroz MV. Analysis of Tc1-Mariner elements in Sclerotinia sclerotiorum suggests recent activity and flexible transposases. BMC Microbiol. 2014a;14: doi: 10.1186/s12866-014-0256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana MF, Silva JCF, Mizubuti ESG, Araújo EF, Condon BJ, Turgeon BG, Queiroz MV. Characterization and potential evolutionary impact of transposable elements in the genome of Cochliobolus heterostrophus . BMC Genomics. 2014b;15: doi: 10.1186/1471-2164-15-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa . Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Selker EU. Repeat-induced gene silencing in fungi. Adv Genet. 2002;46:439–450. doi: 10.1016/s0065-2660(02)46016-6. [DOI] [PubMed] [Google Scholar]
- Shapirova JA. Mobile DNA and evolution in the 21st century. Mob DNA. 2010;1:4–4. doi: 10.1186/1759-8753-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Wilke SK, Ahrén D, Au CH, Birre BW, Borodovsky M, Burns C, Canbäck B, Casselton LA, Cheng CK, et al. From the Cover: Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci U S A. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Huan-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Mogante M, Panaud O, et al. A unified classification system for eukaryotic transposable. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
Internet Resources
- RIPCAL program [accessed March 13, 2014]; // www.sourceforge.net/protects/ripcal/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.