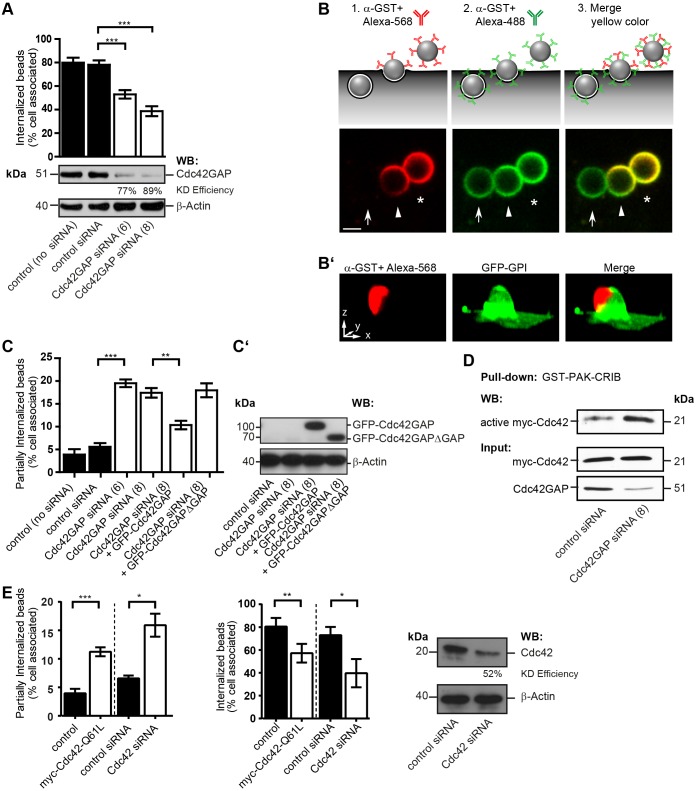
Fig. 2.
Knockdown of Cdc42GAP and Cdc42 hyperactivation inhibit phagocytic cup closure in FnBPA-mediated endothelial cell invasion. (A) HUVECs treated without (control) or with siRNAs specific for luciferase (control siRNA) or Cdc42GAP (siRNAs no. 6 and 8) for 72 h were challenged with FnBPA beads (1-µm diameter) for 30 min and immunostained for extracellular and intracellular beads. Bars represent mean±s.e.m.; 150–200 cells from at least three independent experiments were evaluated. ***P<0.0001 (one-way ANOVA with Bonferroni's post test). A representative western blot (WB) showing knockdown (KD) efficiency of Cdc42GAP siRNAs is also shown. (B) Schematic illustration of the double-staining method to distinguish between extracellular (asterisk), partially internalized (arrowhead) and intracellular (arrow) particles as shown in the lower images. FnBPA beads (3-µm diameter) challenged with HUVECs were immunostained with rabbit anti-GST-antibody and Alexa-Fluor-568-labeled anti-rabbit-IgG antibody (red) before permeabilization, and were then permeabilized and immunostained with the anti-GST-antibody followed by Alexa-Fluor-488-labeled anti-rabbit-IgG antibody (green). Merge of red and green results in yellow color. A yellow cap on a bead (arrowhead) indicates partial internalization. Scale bar: 2 µm. (B′) Three-dimensional reconstruction of partially internalized bead. HUVECs transfected with the plasma membrane marker GFP–GPI (green) were challenged with 3-µm FnBPA beads for 30 min and immunostained for the extracellular portion of the beads using rabbit anti-GST-antibody and Alexa-Fluor-568-labeled secondary antibody (red). The antibody only stained the top part of the bead not enwrapped by plasma membrane giving rise to a cap-like structure. Scale bar: 2.5 µm (correlates with arrow lengths for all dimensions). (C) HUVECs treated without (control) or with siRNAs specific for luciferase (control siRNA) or Cdc42GAP (siRNAs no. 6 and 8) were transfected with vectors expressing either GFP–Cdc42GAP or GFP–Cdc42GAPΔGAP and then challenged with FnBPA beads (1-µm diameter) for 30 min. The percentage of partially internalized beads was determined as described in the Materials and Methods. Results represent mean±s.e.m.; at least 50–200 cells from three independent experiments were evaluated. **P<0.001, ***P<0.0001 (one-way ANOVA with Bonferroni's post test). (C′) Expression of GFP–Cdc42GAP constructs was verified by anti-GFP western blotting. (D) HUVECs treated with control siRNA or Cdc42GAP siRNA no. 8 (similar results were obtained with siRNA no. 6; K.H., data not shown) transfected with a vector for Myc–Cdc42 were subjected to GST–PAK-CRIB pulldown. Bound (active) Myc–Cdc42, the Myc–Cdc42 input and Cdc42GAP knockdown were assessed by western blotting. (E) HUVECs were transfected with empty vector (control) or with a vector for Myc–Cdc42-Q61L, or were treated with siRNAs specific for luciferase (control siRNA) or Cdc42 (no. 17) and then challenged with FnBPA beads (1-µm diameter) for 30 min. The percentage of partially internalized beads (left bar graph) and intracellular beads (right bar graph) was determined. Each bar represents mean±s.e.m. At least 150–200 cells from three independent experiments were evaluated. *P<0.05, **P<0.001, ***P<0.0001 (two-tailed, unpaired Student's t-test). A representative western blot shows the knockdown (KD) efficiency of Cdc42.