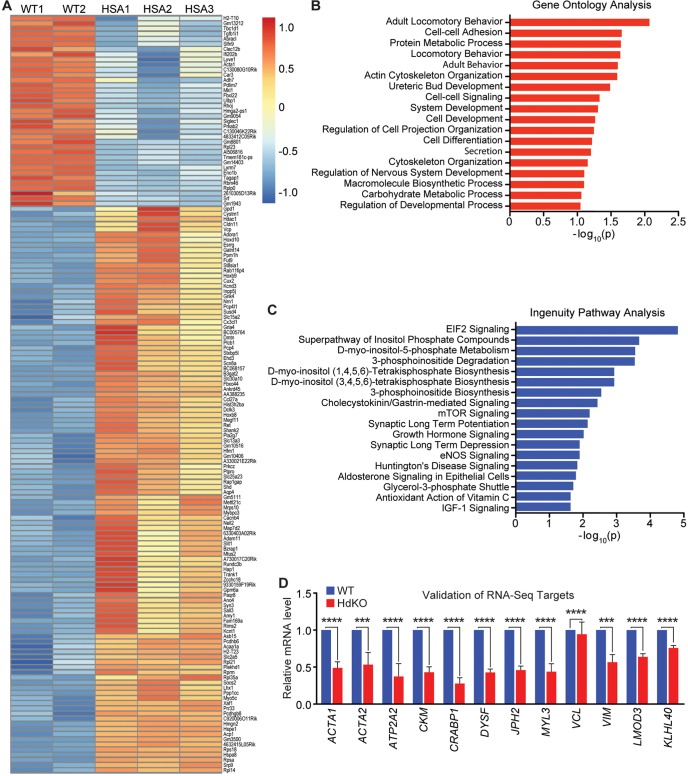
Fig. 6.
MRTF dKO mice demonstrate a dysregulation in cytoskeletal genes. (A) Heat map showing the differentially expressed genes in muscle tissue from WT (MRTF-A+/+; MRTF-B+/+) and MRTF dKO mice at E17.5. (B,C) Gene ontology analysis (B) and ingenuity pathway analysis (IPA) (C) of differentially expressed genes identified top pathways involved in cytoskeletal development and actin cycling. (D) RT-qPCR analysis was performed on E17.5 hindlimb HdKO and WT muscle tissue to validate selected RNA-seq hits and selected MRTF/SRF target genes. Experiments were performed in triplicate with three biological replicates and expression was normalized to 18S rRNA. All data are shown as mean±s.e.m. The Kruskal–Wallis test was used for multiple comparisons, followed by Dunn's post-test for pairwise comparisons. ***P<0.001, ****P<0.0001.